













| General Information: |
|
| Name(s) found: |
JHD2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 728 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEIPALYPT EQEFKNPIDY LSNPHIKRLG VRYGMVKVVP PNGFCPPLSI DMENFTFQPR 60
61 IQNLENLDLK NRCRLFFMKQ LNNFKRSVKD PSKLILREPY TIVEYSDSTH ASEILKKKVY 120
121 FYDVFSELIK DNRTLTDTTQ SFRRKLKFRD ISQLRGDISL WRTISKKFNV PIGLLKEIFE 180
181 KYIASYYIFL HSLNENVHTA LHADQYPKSL LSDDEDDFDL GPDSNSGSDF EEDDDDACIV 240
241 CRKTNDPKRT ILCDSCDKPF HIYCLSPPLE RVPSGDWICN TCIVGNGYYG FTQDTHDYSL 300
301 PEFQEYCKRQ NSRLLPARKL SIDELEEMFW SLVTKNRRSS LTTVKYGADI HNELPGQITG 360
361 FPTREFIPKN INGDELIDYL KYCDHPMNLT NLPMAHNSLL PLFKRNISGM TIPWIYIGSL 420
421 FSTFCWHMED QYTLSANYQH EGDPKVWYSI PESGCTKFND LLNDMSPDLF IKQPDLLHQL 480
481 VTLISPYDPN FKKSGIPVYK AVQKPNEYII TFPKCYHAGF NTGYNFNEAV NFTIDFWLPY 540
541 GFGAITDYKL TQKACVFDMF DLMINVLDKY NKDTLLFNDA FVRQCYSSLI VFYNTELKRI 600
601 RKIQAIVPRT TLLEVHTDPN DEDEEYDIFC SQCKTICSIA FVLRKNNYDS IRTYKRHKKN 660
661 HLSIRQWNEL STTDSKVSIL CTQDYLKSIQ NLNNSDGEEP YIDDELYFTK SLKDIDSLIK 720
721 QVGVKLDR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
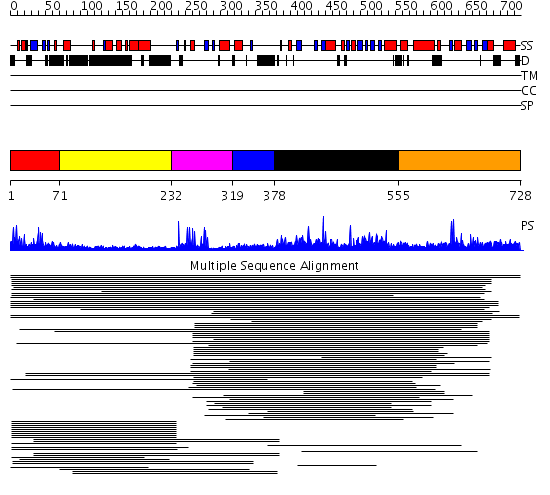
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 25.69897 | jmjN domain No confident structure predictions are available. | |
| 2 | View Details | [71..231] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [232..318] | 98.980454 | Williams-Beuren syndrome transcription factor, WSTF | |
| 4 | View Details | [319..377] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [378..554] | 8.33 | Hypoxia-inducible factor HIF ihhibitor (FIH1) | |
| 6 | View Details | [555..728] | 2.054989 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)