













| General Information: |
|
| Name(s) found: |
IFH1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1085 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGKKSPRKS TINHSTHSGK LPANIKRLIK KGESDTKSRQ SPPTLSTTRP RRFSLIYSSE 60
61 SSLSDVSDSD KNKSTNPHKI KRKAKNISNN SQGKKSKLIQ RQIDNDDEGT ESSDYQAVTD 120
121 GEESENEEEE SEEEEEDDDE DDDDDDDDGS DSDSDSETSS DDENIDFVKL TAQRKKRAMK 180
181 ALSAMNTNSN TLYSSRENSN KNKSVKLSPK KENEEEQKEE KEKEKEEQQK QQESNKKEVN 240
241 GSGTTTTQQA LSFKFKKEDD GISFGNGNEG YNEDIGEEVL DLKNKENNGN EEDKLDSKVM 300
301 LGNNDELRFP NISESDESEY DIDQDAYFDV INNEDSHGEI GTDLETGEDD LPILEEEEQN 360
361 IVSELQNDDE LSFDGSIHEE GSDPVEDAEN KFLQNEYNQE NGYDEEDDEE DEIMSDFDMP 420
421 FYEDPKFANL YYYGDGSEPK LSLSTSLPLM LNDEKLSKLK KKEAKKREQE ERKQRRKLYK 480
481 KTQKPSTRTT SNVDNDEYIF NVFFQSDDEN SGHKSKKGRH KSGKSHIEHK NKGSNLIKSN 540
541 DDLEPSTHST VLNSGKYDSS DDEYDNILLD VAHMPSDDEC SESETSHDAD TDEELRALDS 600
601 DSLDIGTELD DDYEDDDDDS SVTNVFIDID DLDPDSFYFH YDSDGSSSLI SSNSDKENSD 660
661 GSKDCKHDLL ETVVYVDDES TDEDDNLPPP SSRSKNIGSK AKEIVSSNVV GLRPPKLGTW 720
721 ETDNKPFSII DGLSTKSLYA LIQEHQQLRE QHQRAQTPDV KREGSSNGNN GDELTLNELL 780
781 NMSELEDDSP SHTDDMENNY NDAINSKSTN GHAADWYEVP KVPLSAFRNK GINAYEEDEY 840
841 MIPANSNRKV PIGYIGNERT RKKIDKMKEL QRKKTEKKRQ LKKKKKLLKI RKQRQKAIKE 900
901 QETMNLQLGI NGHEIIGNNN SHSDINTGTD FTTNENTPMN ELPSHAPEDA SLIPHNSDLA 960
961 VDSNTRKNST KSVGLDEIHE ILGKDENDLL SVGDINGYDA QEGHVIEDTD ADILASLTAP1020
1021 VQFDNTLSHE NSNSMWRRRQ SMVEAAAENL RFTKNGLFSE SALADIEGIM GNDVNHSFEF1080
1081 NDVLQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
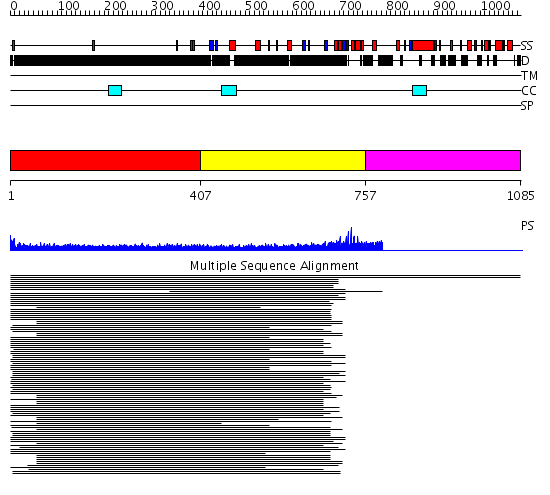
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..406] | 27.045757 | Heavy meromyosin subfragment | |
| 2 | View Details | [407..756] | 7.437997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [757..1085] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)