













| General Information: |
|
| Name(s) found: |
HRK1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 759 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNLLSRNPF HGHHNDHHHD RENSSNNPPQ LIRSSKSFLN FIGRKQSNDS LRSEKSTDSM 60
61 KSTTTTTNYT TTNLNNNTHS HSNATSISTN NYNNNYETNH HHNISHGLHD YTSPASPKQT 120
121 HSMAELKRFF RPSVNKKLSM SQLRSKKHST HSPPPSKSTS TVNLNNHYRA QHPHGFTDHY 180
181 AHTQSAIPPS TDSILSLSNN INIYHDDCIL AQKYGKLGKL LGSGAGGSVK VLVRPTDGAT 240
241 FAVKEFRPRK PNESVKEYAK KCTAEFCIGS TLHHPNVIET VDVFSDSKQN KYYEVMEYCP 300
301 IDFFAVVMTG KMSRGEINCC LKQLTEGVKY LHSMGLAHRD LKLDNCVMTS QGILKLIDFG 360
361 SAVVFRYPFE DGVTMAHGIV GSDPYLAPEV ITSTKSYDPQ CVDIWSIGII YCCMVLKRFP 420
421 WKAPRDSDDN FRLYCMPDDI EHDYVESARH HEELLKERKE KRQRFLNHSD CSAINQQQPA 480
481 HESNLKTVQN QVPNTPASIQ GKSDNKPDIV EEETEENKED DSNNDKESTP DNDKESTIDI 540
541 KISKNENKST VVSANPKKVD ADADADCDAN GDSNGRVDCK ANSDCNDKTD CNANNDCSNE 600
601 SDCNAKVDTN VNTAANANPD MVPQNNPQQQ QQQQQQQQQQ QQQQQQQHHH HQHQNQDKAH 660
661 SIASDNKSSQ QHRGPHHKKI IHGPYRLLRL LPHASRPIMS RILQVDPKKR ATLDDIFNDE 720
721 WFAAIAACTM DSKNKVIRAP GHHHTLVREE NAHLETYKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
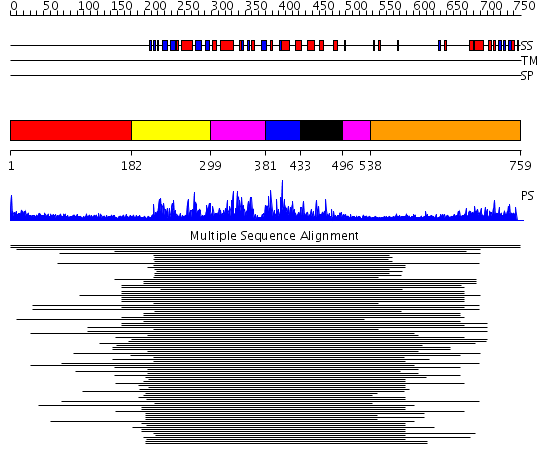
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [182..298] | 295.54902 | Cyclin-dependent PK, CDK2 | |
| 3 | View Details | [299..380] [496..537] |
295.54902 | Cyclin-dependent PK, CDK2 | |
| 4 | View Details | [381..432] | 295.54902 | Cyclin-dependent PK, CDK2 | |
| 5 | View Details | [433..495] | 295.54902 | Cyclin-dependent PK, CDK2 | |
| 6 | View Details | [538..759] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)