













| General Information: |
|
| Name(s) found: |
GCR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 785 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVCTSTSSNF YSIAQYILQS YFKVNVDSLN SLKLVDLIVD QTYPDSLTLR KLNEGATGQP 60
61 YDYFNTVSRD ADISKCPIFA LTIFFVIRWS HPNPPISIEN FTTVPLLDSN FISLNSNPLL 120
121 YIQNQNPNSN SSVKVSRSQT FEPSKELIDL VFPWLSYLKQ DMLLIDRTNY KLYSLCELFE 180
181 FMGRVAIQDL RYLSQHPLLL PNIVTFISKF IPELFQNEEF KGIGSIKNSN NNALNNVTGI 240
241 ETQFLNPSTE EVSQKVDSYF MELSKKLTTE NIRLSQEITQ LKADMNSVGN VCNQILLLQR 300
301 QLLSGNQAIG SKSENIVSST GGGILILDKN SINSNVLSNL VQSIDPNHSK PNGQAQTHQR 360
361 GPKGQSHAQV QSTNSPALAP INMFPSLSNS IQPMLGTLAP QPQDIVQKRK LPLPGSIASA 420
421 ATGSPFSPSP VGESPYSKRF KLDDKPTPSQ TALDSLLTKS ISSPRLPLST LANTAVTESF 480
481 RSPQQFQHSP DFVVGGSSSS TTENNSKKVN EDSPSSSSKL AERPRLPNND STTSMPESPT 540
541 EVAGDDVDRE KPPESSKSEP NDNSPESKDP EKNGKNSNPL GTDADKPVPI SNIHNSTEAA 600
601 NSSGTVTKTA PSFPQSSSKF EIINKKDTKA GPNEAIKYKL SRENKTIWDL YAEWYIGLNG 660
661 KSSIKKLIEN YGWRRWKVSE DSHFFPTRRI IMDYIETECD RGIKLGRFTN PQQPREDIRK 720
721 ILVGDLEKFR INNGLTLNSL SLYFRNLTKN NKEICIFENF KNWNVRSMTE EEKLKYCKRR 780
781 HNTPS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
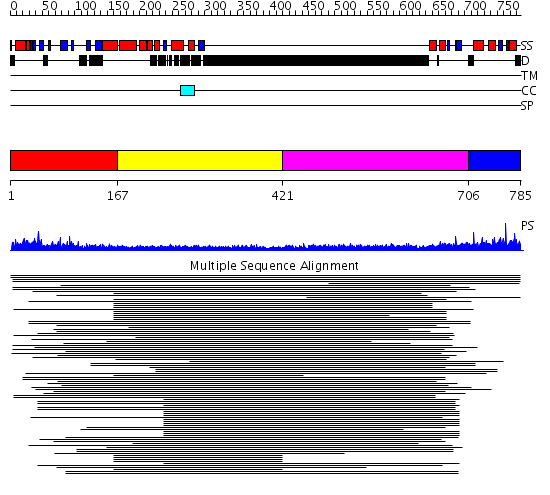
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [167..420] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [421..705] | 3.250996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [706..785] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [674..785] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)