













| General Information: |
|
| Name(s) found: |
FIMB_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIVKLQRKF PILTQEDLFS TIEKFRAIDL DDKGWVEKQQ ALEAVSKDGD ATYDEARETL 60
61 KHVGVDASGR VELDDYVGLV AKLRESKTGA APQTTFNVAP NSTPIVSTAA TGLQHKGKGT 120
121 QAKIIVAGSQ TGTTHTINEE ERREFTKHIN SVLAGDQDIG DLLPFPTDTF QLFDECRDGL 180
181 VLSKLINDSV PDTIDTRVLN WPKKGKELNN FQASENANIV INSAKAIGCV VVNVHSEDII 240
241 EGREHLILGL IWQIIRRGLL SKIDIKLHPE LYRLLEDDET LEQFLRLPPE QILLRWFNYH 300
301 LKQANWNRRV TNFSKDVSDG ENYTILLNQL DPALCSKAPL QTTDLMERAE QVLQNAEKLD 360
361 CRKYLTPSSL VAGNPKLNLA FVAHLFNTHP GLEPIQEEEK PEIEEFDAEG EREARVFTLW 420
421 LNSLDVDPPV ISLFDDLKDG LILLQAYEKV MPGAVDFKHV NKRPASGAEI SRFKALENTN 480
481 YAVDLGRAKG FSLVGIEGSD IVDGNKLLTL GLVWQLMRRN ISITMKTLSS SGRDMSDSQI 540
541 LKWAQDQVTK GGKNSTIRSF KDQALSNAHF LLDVLNGIAP GYVDYDLVTP GNTEEERYAN 600
601 ARLAISIARK LGALIWLVPE DINEVRARLI ITFIASLMTL NK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
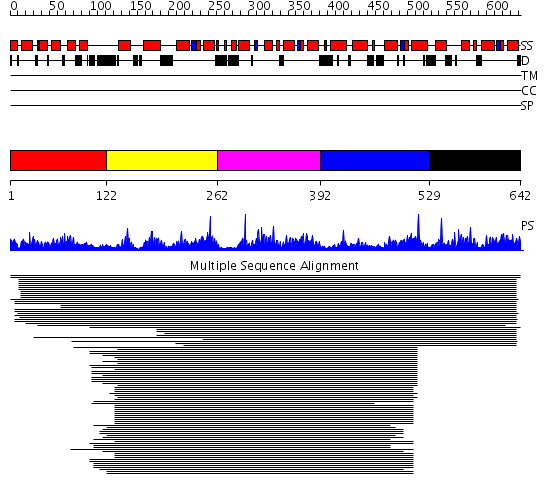
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 45.045757 | Calmodulin | |
| 2 | View Details | [122..261] | 693.450636 | N-terminal actin-crosslinking domain from fimbrin | |
| 3 | View Details | [262..391] | 693.450636 | N-terminal actin-crosslinking domain from fimbrin | |
| 4 | View Details | [392..528] | 6.045757 | Genetically engineered molecular motor | |
| 5 | View Details | [529..642] | 14.99 | beta-spectrin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)