













| General Information: |
|
| Name(s) found: |
FAS2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1887 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKPEVEQELA HILLTELLAY QFASPVRWIE TQDVFLKDFN TERVVEIGPS PTLAGMAQRT 60
61 LKNKYESYDA ALSLHREILC YSKDAKEIYY TPDPSELAAK EEPAKEEAPA PTPAASAPAP 120
121 AAAAPAPVAA AAPAAAAAEI ADEPVKASLL LHVLVAHKLK KSLDSIPMSK TIKDLVGGKS 180
181 TVQNEILGDL GKEFGTTPEK PEETPLEELA ETFQDTFSGA LGKQSSSLLS RLISSKMPGG 240
241 FTITVARKYL QTRWGLPSGR QDGVLLVALS NEPAARLGSE ADAKAFLDSM AQKYASIVGV 300
301 DLSSAASASG AAGAGAAAGA AMIDAGALEE ITKDHKVLAR QQLQVLARYL KMDLDNGERK 360
361 FLKEKDTVAE LQAQLDYLNA ELGEFFVNGV ATSFSRKKAR TFDSSWNWAK QSLLSLYFEI 420
421 IHGVLKNVDR EVVSEAINIM NRSNDALIKF MEYHISNTDE TKGENYQLVK TLGEQLIENC 480
481 KQVLDVDPVY KDVAKPTGPK TAIDKNGNIT YSEEPREKVR KLSQYVQEMA LGGPITKESQ 540
541 PTIEEDLTRV YKAISAQADK QDISSSTRVE FEKLYSDLMK FLESSKEIDP SQTTQLAGMD 600
601 VEDALDKDST KEVASLPNKS TISKTVSSTI PRETIPFLHL RKKTPAGDWK YDRQLSSLFL 660
661 DGLEKAAFNG VTFKDKYVLI TGAGKGSIGA EVLQGLLQGG AKVVVTTSRF SKQVTDYYQS 720
721 IYAKYGAKGS TLIVVPFNQG SKQDVEALIE FIYDTEKNGG LGWDLDAIIP FAAIPEQGIE 780
781 LEHIDSKSEF AHRIMLTNIL RMMGCVKKQK SARGIETRPA QVILPMSPNH GTFGGDGMYS 840
841 ESKLSLETLF NRWHSESWAN QLTVCGAIIG WTRGTGLMSA NNIIAEGIEK MGVRTFSQKE 900
901 MAFNLLGLLT PEVVELCQKS PVMADLNGGL QFVPELKEFT AKLRKELVET SEVRKAVSIE 960
961 TALEHKVVNG NSADAAYAQV EIQPRANIQL DFPELKPYKQ VKQIAPAELE GLLDLERVIV1020
1021 VTGFAEVGPW GSARTRWEME AFGEFSLEGC VEMAWIMGFI SYHNGNLKGR PYTGWVDSKT1080
1081 KEPVDDKDVK AKYETSILEH SGIRLIEPEL FNGYNPEKKE MIQEVIVEED LEPFEASKET1140
1141 AEQFKHQHGD KVDIFEIPET GEYSVKLLKG ATLYIPKALR FDRLVAGQIP TGWNAKTYGI1200
1201 SDDIISQVDP ITLFVLVSVV EAFIASGITD PYEMYKYVHV SEVGNCSGSG MGGVSALRGM1260
1261 FKDRFKDEPV QNDILQESFI NTMSAWVNML LISSSGPIKT PVGACATSVE SVDIGVETIL1320
1321 SGKARICIVG GYDDFQEEGS FEFGNMKATS NTLEEFEHGR TPAEMSRPAT TTRNGFMEAQ1380
1381 GAGIQIIMQA DLALKMGVPI YGIVAMAATA TDKIGRSVPA PGKGILTTAR EHHSSVKYAS1440
1441 PNLNMKYRKR QLVTREAQIK DWVENELEAL KLEAEEIPSE DQNEFLLERT REIHNEAESQ1500
1501 LRAAQQQWGN DFYKRDPRIA PLRGALATYG LTIDDLGVAS FHGTSTKAND KNESATINEM1560
1561 MKHLGRSEGN PVIGVFQKFL TGHPKGAAGA WMMNGALQIL NSGIIPGNRN ADNVDKILEQ1620
1621 FEYVLYPSKT LKTDGVRAVS ITSFGFGQKG GQAIVVHPDY LYGAITEDRY NEYVAKVSAR1680
1681 EKSAYKFFHN GMIYNKLFVS KEHAPYTDEL EEDVYLDPLA RVSKDKKSGS LTFNSKNIQS1740
1741 KDSYINANTI ETAKMIENMT KEKVSNGGVG VDVELITSIN VENDTFIERN FTPQEIEYCS1800
1801 AQPSVQSSFA GTWSAKEAVF KSLGVKSLGG GAALKDIEIV RVNKNAPAVE LHGNAKKAAE1860
1861 EAGVTDVKVS ISHDDLQAVA VAVSTKK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
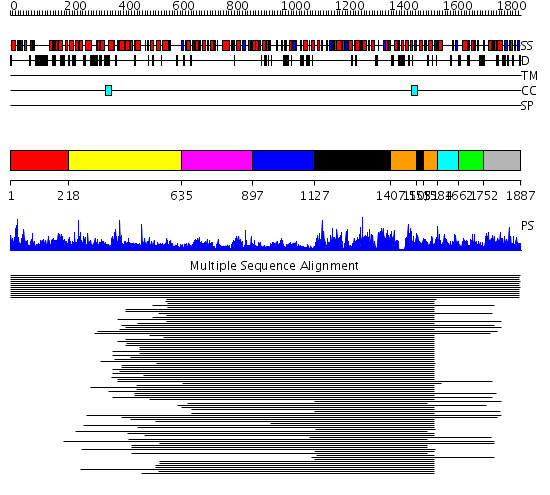
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 1.009936 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [218..634] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [635..896] | 6.221849 | (3R)-hydroxyacyl-CoA dehydrogenase domain of estradiol 17 beta-Dehydrogenase 4 | |
| 4 | View Details | [897..1126] | 3.377997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1127..1406] [1505..1530] |
209.54902 | Beta-ketoacyl-ACP synthase II | |
| 6 | View Details | [1407..1504] [1531..1583] |
209.54902 | Beta-ketoacyl-ACP synthase II | |
| 7 | View Details | [1584..1661] | 5.138995 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1662..1751] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1752..1887] | 5.0 | Holo-(acyl carrier protein) synthase ACPS |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)