













| General Information: |
|
| Name(s) found: |
EST1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 699 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNEEVNEEC MRLFFKNARA HLDKHLTSRL TCDENAYITF RCFLDGIHRK STRFLEELLL 60
61 KQENMYHNNN YERINDSVIP LVLKLLWLQI HEPTLQWFEH WFHDIMRLSN RRKFRVFRIF 120
121 QKKMIQFFKI THRYYYDIIE HLCAKYDMNS VISNALFAKL NLMQYTDGLS THEKIILNTS 180
181 NPLTFSIVIS LQRCVINLGS THFYKTLLNK PSNKPKSVEG FEKSIRYLNI ASLYLPAVGD 240
241 TYFQRAKIYL ITGKFSLYFF ELVRGALVRI PSKCALNNLK DFILTPDFPE RRRLMKKLAI 300
301 LVSKDLKGEK SFFEGQIVLQ FLSIVEHTLV PQSWNASRAS NCWLLKEHLQ MAALKYHSGN 360
361 INVILENLAA TMGSFDLMFT TRKSKEQKNK LKYADLSERQ VFFLDLSFDF IANIIDVVIK 420
421 PSWQKNMEDF RYLAIIRLLM CWIKSYRSIL QYTHRHRKFC TSFALLLNDL INSPLNCSGN 480
481 IYSHRPKRSY LFREDIIFRE FSCINFALTD FNDDYVYDSP DMINNIIGCP TLTKVLSPKE 540
541 ECVLRIRSII FSGMKFLEKN DTGVIWNASK YKFDLISPNI KIKRQIALSE ISSKINVKTQ 600
601 QERVVSSRKV EAKRDEQQRK RAGKIAVTEL EKQFANVRRT KKLSPLPEKD GVSSELVKHA 660
661 ASRGRKTITG PLSSDFLSYP DEAIDADEDI TVQVPDTPT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
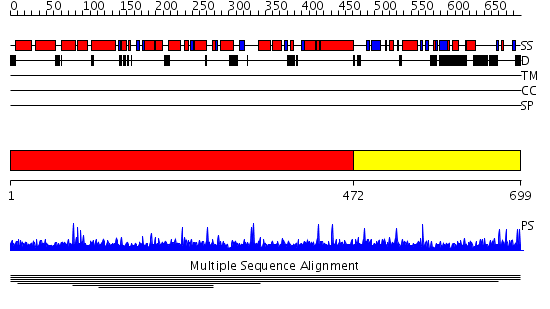
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..471] | 10.41 | Clathrin heavy chain proximal leg segment | |
| 2 | View Details | [472..699] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [286..618] | 2.32 | Crystal structure of the N-terminal domain of human SMG7 | |
| 4 | View Details | [619..699] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)