













| General Information: |
|
| Name(s) found: |
EIF2A_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSQFFLKTS QDIELFQSYP TFEQSNTNSK DFPVISSVLS PCGRFLALST KENVKVFTGP 60
61 CLDNVLLTMK LSDVYDLHFS PAGNYLSTWE RASIQDPNHK NVKVWYLNKP FKKDCVSEDI 120
121 VPAYEYQAKS QSGWFLQFSK LDNYGLRLFK HDLKIVKLSS ANADNFDFQS PFAVLSDDET 180
181 SQHFTTYLIS PAEHPTICTF TPEKGGKPAQ LIIWALSEGK ITKKIASKTF FKADSCQLKW 240
241 NPLGNAILCL AITDFDSSNK SYYGENTLYL LSFQGVNGTL GGNSVRVSLT TGPVHDFTWS 300
301 PTSRQFGVIA GYMPATISFF DLRGNVVHSL PQQAKNTMLF SPSGHYILIA GFGNLQGSVE 360
361 ILDRLDKFKC VSKFDATNTS VCKWSPGGEF IMTATTSPRL RVDNGVKIWH VSGSLVFVKE 420
421 FKELLKVDWR SPCNYKTLEN KDEAFFENHI INNWEPLPDS TTSSLDPKIS NKSELQIHSS 480
481 VQEYISQHPS REASSNGNGS KAKAGGAYKP PHARRTGGGR IVPGVPPGAA KKTIPGLVPG 540
541 MSANKDANTK NRRRRANKKS SETSPDSTPA PSAPASTNAP TNNKETSPEE KKIRSLLKKL 600
601 RAIETLKERQ AVGDKLEDTQ VLKIQTEEKV LKDLEKLGWK DE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
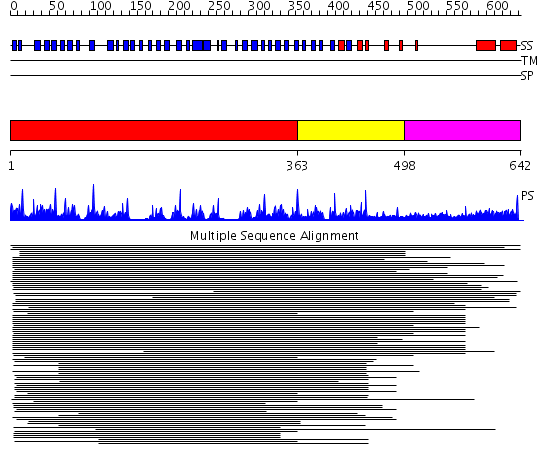
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..362] | 50.0 | Tup1, C-terminal domain | |
| 2 | View Details | [363..497] | 20.30103 | Groucho/tle1, C-terminal domain | |
| 3 | View Details | [498..642] | 3.247996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)