













| General Information: |
|
| Name(s) found: |
EI2BB_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSQAFTSVH PNAATSDVNV TIDTFVAKLK RRQVQGSYAI ALETLQLLMR FISAARWNHV 60
61 NDLIEQIRDL GNSLEKAHPT AFSCGNVIRR ILAVLRDEVE EDTMSTTVTS TSVAEPLISS 120
121 MFNLLQKPEQ PHQNRKNSSG SSSMKTKTDY RQVAIQGIKD LIDEIKNIDE GIQQIAIDLI 180
181 HDHEILLTPT PDSKTVLKFL ITARERSNRT FTVLVTEGFP NNTKNAHEFA KKLAQHNIET 240
241 LVVPDSAVFA LMSRVGKVII GTKAVFVNGG TISSNSGVSS VCECAREFRT PVFAVAGLYK 300
301 LSPLYPFDVE KFVEFGGSQR ILPRMDPRKR LDTVNQITDY VPPENIDIYI TNVGGFNPSF 360
361 IYRIAWDNYK QIDVHLDKNK A |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
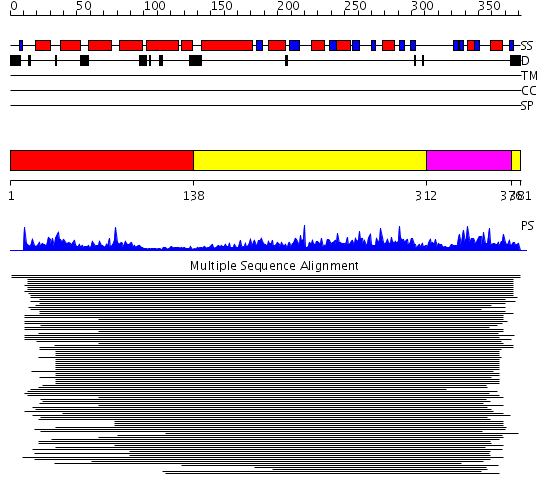
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 1.066988 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [138..311] [376..381] |
10.68 | D-ribose-5-phosphate isomerase (RpiA), catalytic domain; D-ribose-5-phosphate isomerase (RpiA), lid domain | |
| 3 | View Details | [312..375] | 10.68 | D-ribose-5-phosphate isomerase (RpiA), catalytic domain; D-ribose-5-phosphate isomerase (RpiA), lid domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)