













| General Information: |
|
| Name(s) found: |
EF3B_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1044 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSQQSITV LEELFRKLET ATSETREGIS SELSSFLNGN IIEHDVPEVF FDEFQKAIQS 60
61 KQKALNTLGA VAYIANETNL SPSVEPYIVA TVPSVCSKAG SKDNDVQLAA TKALKAIASA 120
121 VNPVAVKALL PHLIHSLETS NKWKEKVAVL EVISVLVDAA KEQIALRMPE LIPVLSESMW 180
181 DTKKGVKEAA TTTITKATET VDNKDIERFI PKLIECIANP NEVPETVHLL GATTFVAEVT 240
241 PATLSIMVPL LSRGLAERET SIKRKAAVII DNMCKLVEDP QVVAPFLGKL LPGLKNNFAT 300
301 IADPEAREVT LKALKTLRRV GNVGEDDVLP EISHAGDVST TLGVIKELLE PEKVAPRFTI 360
361 VVEYIAAIAA NLIDERIIDQ QTWFTHVTPY MTIFLHEKTA KEILDDFRKR AVDNIPVGPN 420
421 FQDEEDEGED LCNCEFSLAY GAKILLNKTQ LRLKRGRRYG LCGPNGAGKS TLMRSIANGQ 480
481 VDGFPTQDEC RTVYVEHDID NTHSDMSVLD FVYSGNVGTK DVITSKLKEF GFSDEMIEMP 540
541 IASLSGGWKM KLALARAVLK DADILLLDEP TNHLDTVNVE WLVNYLNTCG ITSVIVSHDS 600
601 GFLDKVCQYI IHYEGLKLRK YKGNLSEFVQ KCPTAQSYYE LGASDLEFQF PTPGYLEGVK 660
661 TKQKAIVKVS NMTFQYPGTT KPQVSDVTFQ CSLSSRIAVI GPNGAGKSTL INVLTGELLP 720
721 TSGEVYTHEN CRIAYIKQHA FAHIESHLDK TPSEYIQWRF QTGEDRETMD RANRQINEND 780
781 AEAMNKIFKI EGTPRRVAGI HSRRKFKNTY EYECSFLLGE NIGMKSERWV PMMSVDNAWL 840
841 PRGELIESHS KMVAEIDMKE ALASGQFRAL TRKEIELHCA MLGLDSELVS HSRIRGLSGG 900
901 QKVKLVLAAC TWQRPHLIVL DEPTNYLDRD SLGALSKALK AFEGGVIIIT HSAEFTKNLT 960
961 DEVWAVKDGK MTPSGHNWVA GQGAGPRIEK KEEEGDKFDA MGNKINSGKK KSKLSSAELR1020
1021 KKKKERMKKK KEMGDEYVSS DEDF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
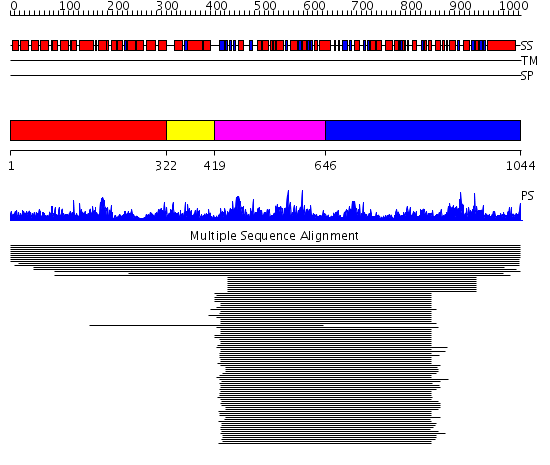
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..321] | 6.11895 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [322..418] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [419..645] | 93.383689 | ATP-binding subunit of the histidine permease | |
| 4 | View Details | [646..1044] | 7.425969 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)