













| General Information: |
|
| Name(s) found: |
DPO4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLKGKFFAF LPNPNTSSNK FFKSILEKKG ATIVSSIQNC LQSSRKEVVI LIEDSFVDSD 60
61 MHLTQKDIFQ REAGLNDVDE FLGKIEQSGI QCVKTSCITK WVQNDKFAFQ KDDLIKFQPS 120
121 IIVISDNADD GQSSTDKESE ISTDVESERN DDSNNKDMIQ ASKPLKRLLQ GDKGRASLVT 180
181 DKTKYKNNEL IIGALKRLTK KYEIEGEKFR ARSYRLAKQS MENCDFNVRS GEEAHTKLRN 240
241 IGPSIAKKIQ VILDTGVLPG LNDSVGLEDK LKYFKNCYGI GSEIAKRWNL LNFESFCVAA 300
301 KKDPEEFVSD WTILFGWSYY DDWLCKMSRN ECFTHLKKVQ KALRGIDPEC QVELQGSYNR 360
361 GYSKCGDIDL LFFKPFCNDT TELAKIMETL CIKLYKDGYI HCFLQLTPNL EKLFLKRIVE 420
421 RFRTAKIVGY GERKRWYSSE IIKKFFMGVK LSPRELEELK EMKNDEGTLL IEEEEEEETK 480
481 LKPIDQYMSL NAKDGNYCRR LDFFCCKWDE LGAGRIHYTG SKEYNRWIRI LAAQKGFKLT 540
541 QHGLFRNNIL LESFNERRIF ELLNLKYAEP EHRNIEWEKK TA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
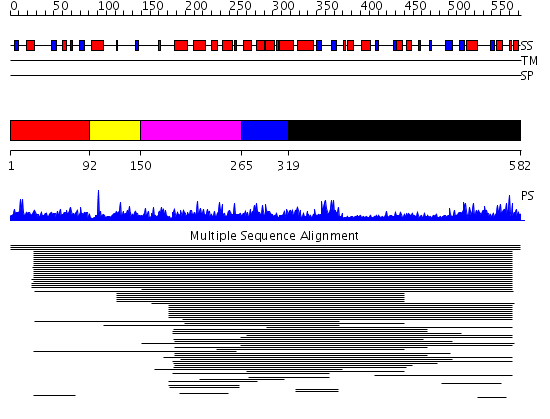
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 1.015993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [92..149] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [150..264] | 216.0103 | Terminal deoxynucleotidyl transferase | |
| 4 | View Details | [265..318] | 216.0103 | Terminal deoxynucleotidyl transferase | |
| 5 | View Details | [319..582] | 216.0103 | Terminal deoxynucleotidyl transferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)