













| General Information: |
|
| Name(s) found: |
DHR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1267 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTYRKRFNE KARSGHMAKL KELKRIRNKQ FTRQDENDER VENPDSAPAE SSTTEPNANA 60
61 EILEPLTEEE KKMKKRKLQE LFTPKESKVS RLKKKRLDKF IEHQLKREER KTIIGKLQDY 120
121 KIDTSLLTSS KRLGEGRQTK KEEFKEALSL ERQGRGNEQT NEILYEEYEP KVWDEYGEGG 180
181 SSEDDDGEDD FEASFGSMPK PTDNEEKKSS GFIDHRPAKF GGSGLSFGFS NIKVINKESK 240
241 TPKKKYNWRQ RVEMEELKKH GKEDEMDFDT TSEDDDEEED QEEEDKMHPS ENPLEEVESA 300
301 DSETGSEKFD QNDVANEFKD WANQEIKKLE GRDQELVTPT LNIDYKPIIR KEDLDDGLQE 360
361 AYVPINENST RKAFYVEVSR SDEIQKARIQ LPVFGEEHKI MEAIHHNDVV IICGETGSGK 420
421 TTQVPQFLYE AGFGAEDSPD YPGMVGITQP RRVAAVSMAE RVANELGDHG HKVGYQIRFD 480
481 STAKEDTKVK FMTDGVLLRE MMHDFKLTKY SSIIIDEAHE RNINTDILIG MLSRCVRLRA 540
541 KLHKENPIEH KKLKLIIMSA TLRVSDFSEN KTLFPIAPPV LQVDARQFPV SIHFNRRTAF 600
601 NYTDEAFRKT CKIHQKLPPG AILVFLTGQQ EITHMVKRLR KEFPFKKNSK YNKDLETPVS 660
661 KMGINSKTTD LEAEDIDFSV QVIDQDKFKS AIRYEEDEGN SGNGEDEEDE EEEGFEEVLT 720
721 EGQTANDPLY VLPLYSLLPT KEQMRVFQKP PQGSRLCIVA TNVAETSLTI PGVRYVVDSG 780
781 RSKERKYNES NGVQSFEVGW VSKASANQRS GRAGRTGPGH CYRLYSSAVF EHDFEQFSKP 840
841 EILRMPVESI VLQMKSMAIH NIINFPFPTP PDRVALSKAI QLLQYLGALD NKEMITEDGK 900
901 KMSLFPLSPR FSKMLLVSDE KACLPYIVAI VSALSVGDPF INEFELGINE ISRKPNPDEN 960
961 LDDKIREHDE STPGMDPELK KELRSKFYKS RSQFSKLDKF SDVFRLLSVV SAMDYVPKEQ1020
1021 KEIFMKKNFL RGKLMEEIVK LRKQLMYIIK SNTSKENIAV VIRNEDLKSD IPSVIQIKLL1080
1081 KQMICAGFVD HVAVRADVLF PDDAKITNRT SIINIPYIPV LATRTPNIED CFVYIHPTSI1140
1141 LNNLGEMPPK YMLYYSLHLG GNNKTRMNTL CDIASTPLAN IARKGLLLTY SKPLTGQGLK1200
1201 TVNLSPTERY CYVVPRFGST VDNDLKIGWD LNPIAVHQKK QKGQWTVIKF ITRKGFQTIT1260
1261 GEEKEKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
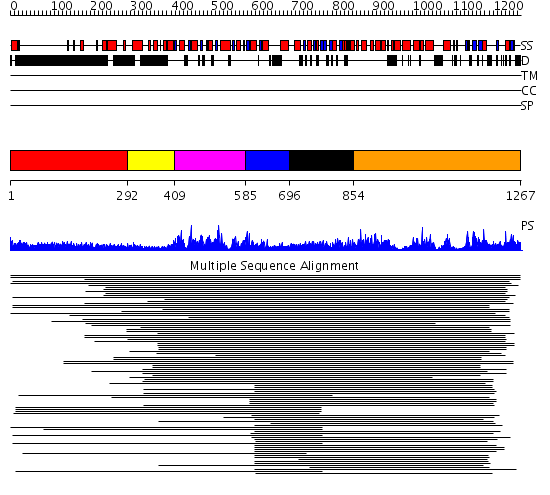
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | 47.30103 | Heavy meromyosin subfragment | |
| 2 | View Details | [292..408] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [409..584] | 4.221849 | Putative DEAD box RNA helicase | |
| 4 | View Details | [585..695] | 4.221849 | Putative DEAD box RNA helicase | |
| 5 | View Details | [696..853] | 12.3 | Initiation factor 4a | |
| 6 | View Details | [854..1267] | 4.132947 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)