













| General Information: |
|
| Name(s) found: |
DCR2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRLPRLYQR YLLYLVVFVV IALFYFLQAP RVEEHIGFDL ALPISHVDNL WFQNKGLEGF 60
61 SNDDKLVVNI GYDECFHIGR FYEGCFNRHE LKSTLTDGHQ YLQRKRIHKD LRGSFGRRWF 120
121 GKSEYLYYDV LYPALVDYFG SNLEKLNVEA VTGISKYPKD KSLPFMDVSI TFEPISIELL 180
181 QKRSYISDIN ILFGVDCIQP IANWTLQKEF PLVKYRYSEP AYLTYKFVGT RPVDTGAQRL 240
241 QETDEGKFKI VQLADLHLGV GESECIDEYP KHEACKADPK TETFVQQVLD IEKPQLVVFT 300
301 GDQIMGDRSI QDSETVLLKA VAPVIARKIP WAMVWGNHDD EGSLTRWQLS EIASVLPYSL 360
361 FKFSPHDTHD NTFGVGNYIY QIFSNNDTEV PVGTLYFLDS HKYSTVGKIY PGYDWIKESQ 420
421 WKYIEDYHDV NLKFKTGLSM AFFHIPLPEY LNIESKTHPG EKNPLIGMYK EGVTAPKYNS 480
481 EGITTLDRLS VDVVSCGHDH CNDYCLRDDS TPNKIWLCYG GGGGEGGYAG YGGTERRIRI 540
541 YEINVNENNI HTWKRLNGSP KEIFDFQSML DGNSPESV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
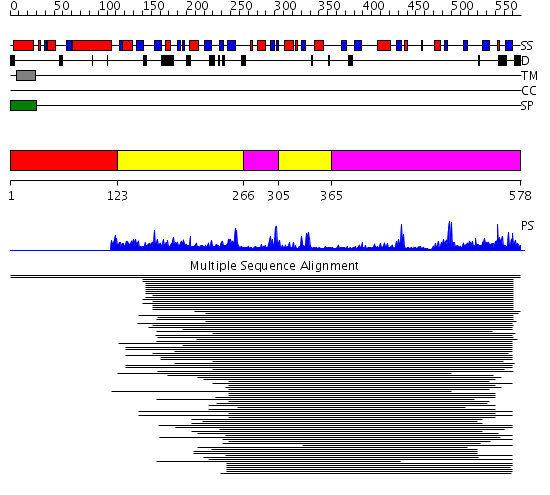
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [123..265] [305..364] |
68.69897 | Purple acid phosphatase, N-terminal domain; Plant purple acid phosphatase, catalytic domain | |
| 3 | View Details | [266..304] [365..578] |
68.69897 | Purple acid phosphatase, N-terminal domain; Plant purple acid phosphatase, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|