













| General Information: |
|
| Name(s) found: |
COG6_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 839 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFVVDYQTY AMADTATPEL PEPEPRLNLT SDAQSQPTGK LDLQFKLPDL QRYSNNNATL 60
61 PVDNDGAGSK DLHKKMTHYA MSSIDKIQLS NPSKQLGQNS QDEKLSQQES QNFTNYEPKN 120
121 LDLSKLVSPS SGSNKNTTNL VLSNKLSKIL NNYTLINYQA TVQLRKSLKV LEENKERLSL 180
181 DEQKLMNPEY VGTLARRALR TDLESQLLKE HITVLEEFKP IIRRIKRLSS SVEKIQRTSE 240
241 KLLSNETNEV PTNNVVLQEI DQYRLKAEQL KLKKKILLSI RDRFTLNQVE DDVITNGTID 300
301 NIFFEVVKKV INIKDESSFL LTLPNLNAGN ALIMGVNEIL EKTNKKIFNY LIDFLYSFES 360
361 SSNLLNDHGT TEQESLNIFR KSLVFLSSDL ELFNELLKRV TTLRSKSILD EFLSQFDMNS 420
421 TTSKPIILSA HDPIRYIGDV LASVHSIIAN EADFVKSLFD FQDEDLKDTP ISILQQNKTF 480
481 LKGIDNKLLN DIIQSLSNSC RIRIEQIVRF EENPIINFEI VRLLKLYRVM FERKGIQDDS 540
541 SIINNLKSLE DISKNRIIGY YEDYMKQTVM AETKNSSDDL LPPEWLSEYM NKLVELFEIY 600
601 EKTHAAEDEE SEDNKLLSSK NLQTIVEQPI KDVLLKQLQT SFPLAKKNEK EKASLLTIEI 660
661 NCFDLIKSRL QPFEGLFAQD DDSRKITIWV CDKLKEYTKQ MLTLQIKFLF ENTGLDLYSN 720
721 LVNMIFPVDS VKDELDYDMY LALRDNSLME LDMVRKNVHD KLNYYLPQAL TDVQGNLLFK 780
781 LTSPMIADEI CDECFKKLSL FYNIFRKLLI HLYPNKKDQV FEILNFSTDE FDMLIGIDH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
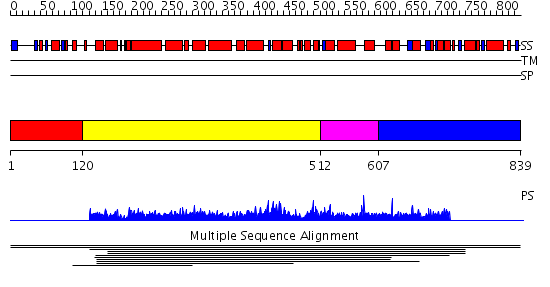
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [120..511] | 1.0079 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [512..606] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [607..839] | 9.12 | (+)-bornyl diphosphate synthase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)