













| General Information: |
|
| Name(s) found: |
CIN1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1014 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNIRALLDS IQSGVQTVSP EKHQQTIAAI NKFQDDPALL DTILPRCVPL LTKSFFCMSQ 60
61 RDQKLVAELF YNLDKISHSK VLKSLDTSIF RLNEILNYLQ DRASPSSFSD VLCVYLNLSW 120
121 LSVILLSPYA FKDKFNKTLQ VSSRFENYPI CIPPINKIKA VLYFKNFTDA FDQLPEREQA 180
181 NVPFLNQFLK LFIQSSEKAN YYFSNENLRH LQQVALSNDG IKLLPKLFQI SFNHGSHDIL 240
241 DAIVEFFHDH LSSNSTDTRF QLAHSFAKIA KFLHQADPAS FIELIDYTIE NTVSLLQAPC 300
301 DSIDSNELHT SLLIIAEVAL AKILPIDLVD RVLTLIIPKT CHFQKSHFQI IKGHHIRDST 360
361 NFIIWSVIRS NRSNSLSPQV LQSLLSHLLI NAFFDPELII RYSSFAALQE LLGRSNKSLA 420
421 LNQNDIALIL QANWKDLPRS FEENSGLIRR LFNPENTSKS AVCVWKVFRD WSFNWNLLEN 480
481 LHLTTMKLNI DYNLVPLIKS KLSSPALLQE VLNKAGSSVT QNCQILYLYL KLFENDVNCP 540
541 KISEICIDIY QKKIKFQLTT QAKRQFNDNS PELFQIFVIL KYWQLTGQND FNQELFWKFV 600
601 DIVSPQKKLN LYNEFIPIIQ QIISQCVSLN YTRIVQLIKS DNELTCRSIC HMPDQEKMCS 660
661 LFFSQFPLLS PQSRSLLIGE LDHHWDVRIS LLPSNSYRKF RNIIINCLDD YTITQQGDVG 720
721 RLVRIQALKL MQSHPDFLSG DCDSINPKLT RLLAEPVPEI RKLSYQLLAS ATSQITVLSD 780
781 SSILNFRHKQ GLSEEFWKGY AVSAGAIHFT DSQLTSSIDS FIVYYRSLSP SQQLELCHDL 840
841 IRIIPSAKQI AESRIRDRNK DPLTGGMRFD TIKFTIHCVK FWTRIMESGL VVLHPNFNFQ 900
901 GVFAKFYNLH LLDCTTLRVS VIKFFPFLAI SCYHTMRENA DQKNLSNIIL KRLLVLVKRE 960
961 YAATKSKFMT DQNVALQGMF QIFLELGVTR QLQALQVACQ KHELANILES DITL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
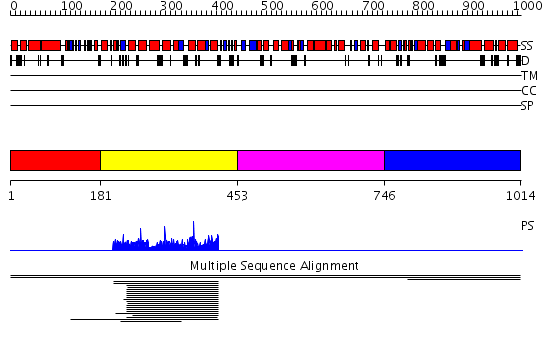
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..180] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [181..452] | 11.013952 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [453..745] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [746..1014] | 1.00287 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)