













| General Information: |
|
| Name(s) found: |
CFT2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTYKYNCCDD GSGTTVGSVV RFDNVTLLID PGWNPSKVSY EQCIKYWEKV IPEIDVIILS 60
61 QPTIECLGAH SLLYYNFTSH FISRIQVYAT LPVINLGRVS TIDSYASAGV IGPYDTNKLD 120
121 LEDIEISFDH IVPLKYSQLV DLRSRYDGLT LLAYNAGVCP GGSIWCISTY SEKLVYAKRW 180
181 NHTRDNILNA ASILDATGKP LSTLMRPSAI ITTLDRFGSS QPFKKRSKIF KDTLKKGLSS 240
241 DGSVIIPVDM SGKFLDLFTQ VHELLFESTK INAHTQVPVL ILSYARGRTL TYAKSMLEWL 300
301 SPSLLKTWEN RNNTSPFEIG SRIKIIAPNE LSKYPGSKIC FVSEVGALIN EVIIKVGNSE 360
361 KTTLILTKPS FECASSLDKI LEIVEQDERN WKTFPEDGKS FLCDNYISID TIKEEPLSKE 420
421 ETEAFKVQLK EKKRDRNKKI LLVKRESKKL ANGNAIIDDT NGERAMRNQD ILVENVNGVP 480
481 PIDHIMGGDE DDDEEEENDN LLNLLKDNSE KSAAKKNTEV PVDIIIQPSA ASKHKMFPFN 540
541 PAKIKKDDYG TVVDFTMFLP DDSDNVNQNS RKRPLKDGAK TTSPVNEEDN KNEEEDGYNM 600
601 SDPISKRSKH RASRYSGFSG TGEAENFDNL DYLKIDKTLS KRTISTVNVQ LKCSVVILNL 660
661 QSLVDQRSAS IIWPSLKSRK IVLSAPKQIQ NEEITAKLIK KNIEVVNMPL NKIVEFSTTI 720
721 KTLDISIDSN LDNLLKWQRI SDSYTVATVV GRLVKESLPQ VNNHQKTASR SKLVLKPLHG 780
781 SSRSHKTGAL SIGDVRLAQL KKLLTEKNYI AEFKGEGTLV INEKVAVRKI NDAETIIDGT 840
841 PSELFDTVKK LVTDMLAKI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
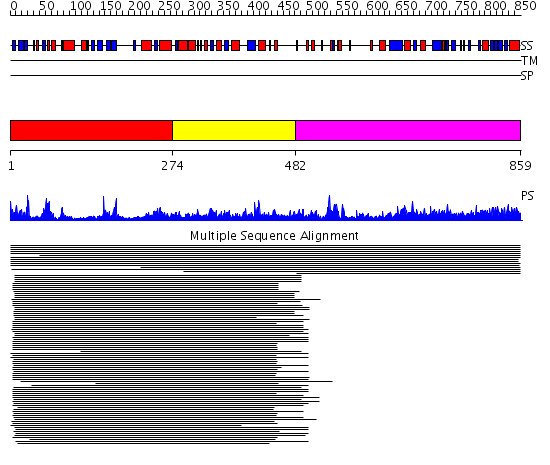
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..273] | 10.69897 | Rubredoxin oxygen:oxidoreductase (ROO), C-terminal domain; Rubredoxin oxygen:oxidoreductase (ROO), N-terminal domain | |
| 2 | View Details | [274..481] | 1.17196 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [482..859] | 1.05888 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)