













| General Information: |
|
| Name(s) found: |
CDC24_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 854 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIQTRFASG TSLSDLKPKP SATSISIPMQ NVMNKPVTEQ DSLFHICANI RKRLEVLPQL 60
61 KPFLQLAYQS SEVLSERQSL LLSQKQHQEL LKSNGANRDS SDLAPTLRSS SISTATSLMS 120
121 MEGISYTNSN PSATPNMEDT LLTFSMGILP ITMDCDPVTQ LSQLFQQGAP LCILFNSVKP 180
181 QFKLPVIASD DLKVCKKSIY DFILGCKKHF AFNDEELFTI SDVFANSTSQ LVKVLEVVET 240
241 LMNSSPTIFP SKSKTQQIMN AENQHRHQPQ QSSKKHNEYV KIIKEFVATE RKYVHDLEIL 300
301 DKYRQQLLDS NLITSEELYM LFPNLGDAID FQRRFLISLE INALVEPSKQ RIGALFMHSK 360
361 HFFKLYEPWS IGQNAAIEFL SSTLHKMRVD ESQRFIINNK LELQSFLYKP VQRLCRYPLL 420
421 VKELLAESSD DNNTKELEAA LDISKNIARS INENQRRTEN HQVVKKLYGR VVNWKGYRIS 480
481 KFGELLYFDK VFISTTNSSS EPEREFEVYL FEKIIILFSE VVTKKSASSL ILKKKSSTSA 540
541 SISASNITDN NGSPHHSYHK RHSNSSSSNN IHLSSSSAAA IIHSSTNSSD NNSNNSSSSS 600
601 LFKLSANEPK LDLRGRIMIM NLNQIIPQNN RSLNITWESI KEQGNFLLKF KNEETRDNWS 660
661 SCLQQLIHDL KNEQFKARHH SSTSTTSSTA KSSSMMSPTT TMNTPNHHNS RQTHDSMASF 720
721 SSSHMKRVSD VLPKRRTTSS SFESEIKSIS ENFKNSIPES SILFRISYNN NSNNTSSSEI 780
781 FTLLVEKVWN FDDLIMAINS KISNTHNNNI SPITKIKYQD EDGDFVVLGS DEDWNVAKEM 840
841 LAENNEKFLN IRLY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
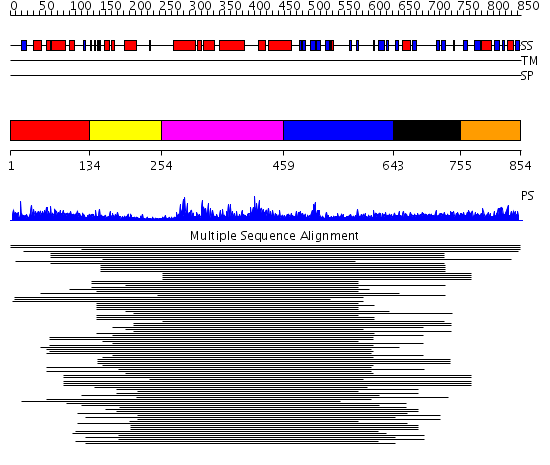
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 1.108997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..253] | 11.69897 | Calponin | |
| 3 | View Details | [254..458] | 202.457575 | GEF of TIAM1 (T-Lymphoma invasion and metastasis indusing protein 1) | |
| 4 | View Details | [459..642] | 202.457575 | GEF of TIAM1 (T-Lymphoma invasion and metastasis indusing protein 1) | |
| 5 | View Details | [643..754] | 1.061996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [755..854] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)