













| General Information: |
|
| Name(s) found: |
ARP8_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 881 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQEEAESSI IYEEPIDIPL EDDDDEDELE EENSVPLSSQ ADQENAENES DDSVDNVVGS 60
61 ETPRSVTGLS VDPRDVADEE DEDEEGEDED EDEDDNDVDN EDENDNDNAN ENENELGSSR 120
121 DKRAPPAVQT SKRYKKYPKL DPAKAPPGKK VPLHLLEKRR LGRIKAAEEF AKTLKKIGIE 180
181 KVETTTLPAT GLFQPLMLIN QKNYSSDYLK KDDQIFALRD RKFLRNNNTS QISSTNTPDV 240
241 IDLKSLPHSE ASAAPLNDEI DLNDPTATIV IHPGSNSIKI GFPKDDHPVV VPNCVAVPKK 300
301 WLDLENSEHV ENVCLQREQS EEFNNIKSEM EKNFRERMRY YKRKVPGNAH EQVVSFNENS 360
361 KPEIISEKND PSPIEWIFDD SKLYYGSDAL RCVDEKFVIR KPFRGGSFNV KSPYYKSLAE 420
421 LISDVTKLLE HALNSETLNV KPTKFNQYKV VLVIPDIFKK SHVETFIRVL LTELQFQAVA 480
481 IIQESLATCY GAGISTSTCV VNIGAAETRI ACVDEGTVLE HSAITLDYGG DDITRLFALF 540
541 LLQSDFPLQD WKIDSKHGWL LAERLKKNFT TFQDADVAVQ LYNFMNRSPN QPTEKYEFKL 600
601 FDEVMLAPLA LFFPQIFKLI RTSSHKNSSL EFQLPESRDL FTNELNDWNS LSQFESKEGN 660
661 LYCDLNDDLK ILNRILDAHN IIDQLQDKPE NYGNTLKENF APLEKAIVQS IANASITADV 720
721 TRMNSFYSNI LIVGGSSKIP ALDFILTDRI NIWRPSLLSS ASFPQFYKKL TKEIKDLEGH 780
781 YVNAPDKTED ENKQILQAQI KEKIVEELEE QHQNIEHQNG NEHIFPVSII PPPRDMNPAL 840
841 IIWKGASVLA QIKLVEELFI TNSDWDVHGS RILQYKCIFT Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
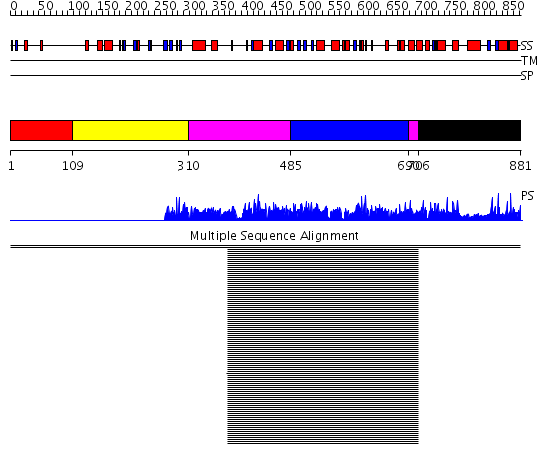
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 3.005997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [109..309] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [310..484] [690..705] |
1023.9794 | Actin | |
| 4 | View Details | [485..689] | 1023.9794 | Actin | |
| 5 | View Details | [706..881] | 13.93 | Actin-related protein 3, Arp3 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)