













| General Information: |
|
| Name(s) found: |
ARO1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1588 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQLAKVPIL GNDIIHVGYN IHDHLVETII KHCPSSTYVI CNDTNLSKVP YYQQLVLEFK 60
61 ASLPEGSRLL TYVVKPGETS KSRETKAQLE DYLLVEGCTR DTVMVAIGGG VIGDMIGFVA 120
121 STFMRGVRVV QVPTSLLAMV DSSIGGKTAI DTPLGKNFIG AFWQPKFVLV DIKWLETLAK 180
181 REFINGMAEV IKTACIWNAD EFTRLESNAS LFLNVVNGAK NVKVTNQLTN EIDEISNTDI 240
241 EAMLDHTYKL VLESIKVKAE VVSSDERESS LRNLLNFGHS IGHAYEAILT PQALHGECVS 300
301 IGMVKEAELS RYFGILSPTQ VARLSKILVA YGLPVSPDEK WFKELTLHKK TPLDILLKKM 360
361 SIDKKNEGSK KKVVILESIG KCYGDSAQFV SDEDLRFILT DETLVYPFKD IPADQQKVVI 420
421 PPGSKSISNR ALILAALGEG QCKIKNLLHS DDTKHMLTAV HELKGATISW EDNGETVVVE 480
481 GHGGSTLSAC ADPLYLGNAG TASRFLTSLA ALVNSTSSQK YIVLTGNARM QQRPIAPLVD 540
541 SLRANGTKIE YLNNEGSLPI KVYTDSVFKG GRIELAATVS SQYVSSILMC APYAEEPVTL 600
601 ALVGGKPISK LYVDMTIKMM EKFGINVETS TTEPYTYYIP KGHYINPSEY VIESDASSAT 660
661 YPLAFAAMTG TTVTVPNIGF ESLQGDARFA RDVLKPMGCK ITQTATSTTV SGPPVGTLKP 720
721 LKHVDMEPMT DAFLTACVVA AISHDSDPNS ANTTTIEGIA NQRVKECNRI LAMATELAKF 780
781 GVKTTELPDG IQVHGLNSIK DLKVPSDSSG PVGVCTYDDH RVAMSFSLLA GMVNSQNERD 840
841 EVANPVRILE RHCTGKTWPG WWDVLHSELG AKLDGAEPLE CTSKKNSKKS VVIIGMRAAG 900
901 KTTISKWCAS ALGYKLVDLD ELFEQQHNNQ SVKQFVVENG WEKFREEETR IFKEVIQNYG 960
961 DDGYVFSTGG GIVESAESRK ALKDFASSGG YVLHLHRDIE ETIVFLQSDP SRPAYVEEIR1020
1021 EVWNRREGWY KECSNFSFFA PHCSAEAEFQ ALRRSFSKYI ATITGVREIE IPSGRSAFVC1080
1081 LTFDDLTEQT ENLTPICYGC EAVEVRVDHL ANYSADFVSK QLSILRKATD SIPIIFTVRT1140
1141 MKQGGNFPDE EFKTLRELYD IALKNGVEFL DLELTLPTDI QYEVINKRGN TKIIGSHHDF1200
1201 QGLYSWDDAE WENRFNQALT LDVDVVKFVG TAVNFEDNLR LEHFRDTHKN KPLIAVNMTS1260
1261 KGSISRVLNN VLTPVTSDLL PNSAAPGQLT VAQINKMYTS MGGIEPKELF VVGKPIGHSR1320
1321 SPILHNTGYE ILGLPHKFDK FETESAQLVK EKLLDGNKNF GGAAVTIPLK LDIMQYMDEL1380
1381 TDAAKVIGAV NTVIPLGNKK FKGDNTDWLG IRNALINNGV PEYVGHTAGL VIGAGGTSRA1440
1441 ALYALHSLGC KKIFIINRTT SKLKPLIESL PSEFNIIGIE STKSIEEIKE HVGVAVSCVP1500
1501 ADKPLDDELL SKLERFLVKG AHAAFVPTLL EAAYKPSVTP VMTISQDKYQ WHVVPGSQML1560
1561 VHQGVAQFEK WTGFKGPFKA IFDAVTKE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
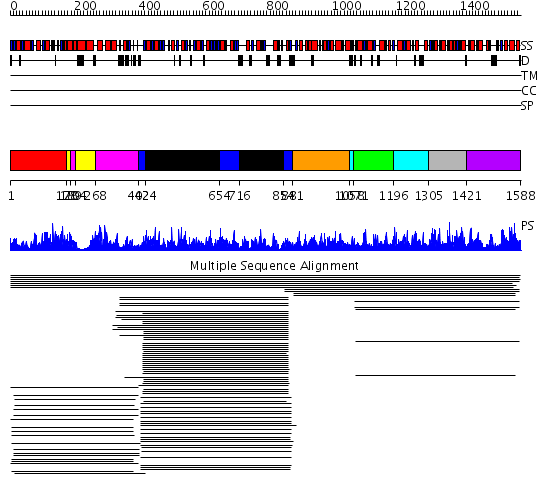
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | 1193.0 | Dehydroquinate synthase, DHQS | |
| 2 | View Details | [178..188] [204..267] |
1193.0 | Dehydroquinate synthase, DHQS | |
| 3 | View Details | [189..203] [268..401] |
1193.0 | Dehydroquinate synthase, DHQS | |
| 4 | View Details | [402..423] [654..715] [854..880] |
788.9897 | No description for 1ln5A_ was found. | |
| 5 | View Details | [424..653] [716..853] |
788.9897 | No description for 1ln5A_ was found. | |
| 6 | View Details | [881..1057] | 166.69897 | Shikimate kinase (AroK) | |
| 7 | View Details | [1058..1070] [1196..1304] |
149.07631 | Type I 3-dehydroquinate dehydratase | |
| 8 | View Details | [1071..1195] | 149.07631 | Type I 3-dehydroquinate dehydratase | |
| 9 | View Details | [1305..1420] | 5.30103 | Shikimate / quinate 5-dehydrogenase No confident structure predictions are available. | |
| 10 | View Details | [1421..1588] | 8.221849 | Glutamyl tRNA-reductase dimerization domain; Glutamyl tRNA-reductase middle domain; Glutamyl tRNA-reductase catalytic, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)