













| General Information: |
|
| Name(s) found: |
ALK2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 676 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFDAVADQQ MTDRRYFALE VAESDDADSS LNSSSMGSPA VDVGRKVYKI TSHKGSAEDE 60
61 SQSFFTSSDS PTSKTRPVGK TIENDDYYGK RSSTGSSLKQ LFNKININDT AHSSNKENVS 120
121 QSVLSENKLL SPSKRLSKQG LTKVTNSKFR TPLRPISNQS TLSRDEPVKD FRSLKFRSGS 180
181 DFKCWGDEKT SSHVHSSSVN SVNSFTSTTS SSKWKFWKND NLLSRSLSSR SVNDQDPNFV 240
241 QPKPTNSLQK KSSISSFHNS IFGGGKHTEK KRNSGFIMPD HQSTKELNHK HSSSNLSFRS 300
301 LKHKTSHSSL NKLKVRRKGN TQELNHPIKK TCQISLPVPD QVSKDKIQLK LKNSTSLASL 360
361 SSEVTPINTL DYNDSILQQI LQLCDVKYIL HDLREAQSLG LFTLNTRSVQ LSHNFWQTYH 420
421 SDMQTSLICK KVCLGALSDL TTSNLISLHE LKSLRLIQGT SGVANLLQAY VVPSNQCEND 480
481 QNLILYLFFK YQGTPLSRCS NIDYSQALSI FWQCSSILYV AESKFQLEHR NLTLDHILID 540
541 SKGNVTLIDM KCCRFLNIDN NKASYTRLDH HYFFQGRGTL QFEIYELMRS MLPQPISWAT 600
601 FEPRTNLLWL YHLSSSLLKM AKKAVVSGAL NREENILIEL THLLDPARKH SKTIFKKELV 660
661 IRTCGDLLSL KGEIMQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
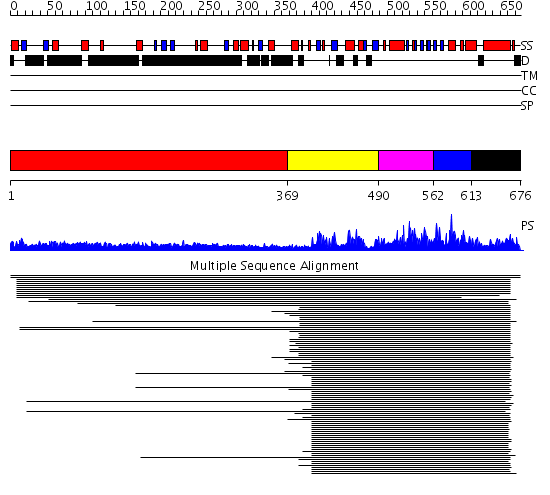
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..368] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [369..489] | 101.0 | Cyclin-dependent PK, CDK2 | |
| 3 | View Details | [490..561] | 101.0 | Cyclin-dependent PK, CDK2 | |
| 4 | View Details | [562..612] | 101.0 | Cyclin-dependent PK, CDK2 | |
| 5 | View Details | [613..676] | 101.0 | Cyclin-dependent PK, CDK2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)