













| General Information: |
|
| Name(s) found: |
SAC4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 831 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSSPSVENG GSGSSGSSAL LGCMQQFKLF ETQANFYMIG WNGSGVYRIL KIDRLEASEL 60
61 NLREDSTAYT KKECYELLKR IHEGNKATGG LKLVTVCYGI IGFIKFLGPY YMLLITERRE 120
121 IGEICGHIVY EVSKSDMIAL QHSSVLCNTA NLRDENRYKR LLCMVDLTKD FFFSYSYNIM 180
181 RSFQKNICDH ESGGTLYKKM FVWNEFLTRG TRHHLRNTLW TVALVYGFFK QTILSEAGRN 240
241 FKLTLIARRS RHNAGTRYLK RGINESGNVA NDVETEQIVS EDVPVDRPMQ ISSVVQNRGS 300
301 IPLFWSQETS RMKVKPDIVL SKRDLNYEAT RVHFENLVER YGVPIIILNL IKTNERKPRE 360
361 SILRAEFANA IDFINKDLPE ENRLRFLHWD LHKHFHSKTE NVLALLGKVA ACALMLTGFF 420
421 YYQLTPAMKL EGYMSLSSSD ADTSPHNSSD DDSRDYDSLE KNCRPSKNVA NGDYDVKPSR 480
481 LQSGVLRTNC IDCLDRTNVA QYAYGWAALG QQLHALGIRD APTIELDDPL SSTLMGLYER 540
541 MGDTLAYQYG GSAAHNKVFS ERRGQWRAAT QSQEFLRTLQ RYYNNAYMDA DKQDAINIFL 600
601 GTFRPEQGSQ AVWELRSDSH SNGRSGEISM GEDEKFLVKR CLSDGNILHE SHTPMSAMSR 660
661 KNESISHRGF VSSHQVTRTH IISESSPDMP AAGDVTLSRC TPSMPSTHFF GDVQKVQHNG 720
721 SSSIYLSEQE DMSSVSNFVD IEWLSSSENL CENDHLSRPS ALTIYSTAET SSSENIITEV 780
781 KQLTPAMRES GSSSRKGKEP VETELSVHTK IRDDFPDSFK QWVAYGEALC H |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
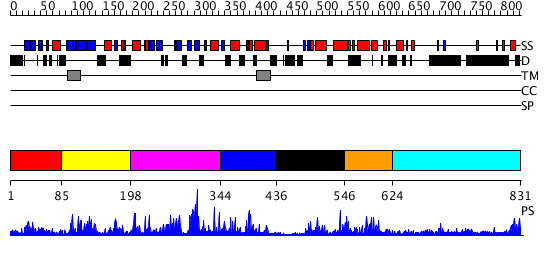
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..197] | 4.180989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [198..343] | 1.191979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [344..435] | 2.19599 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [436..545] | 1.19799 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [546..623] | 3.186993 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [624..831] | 1.131996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)