













| General Information: |
|
| Name(s) found: |
SSDH_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 528 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrial matrix [IDA] mitochondrion [IDA] |
| Biological Process: |
response to heat
[IMP]
oxygen and reactive oxygen species metabolic process [IMP] gamma-aminobutyric acid catabolic process [IDA] glutamate decarboxylation to succinate [IDA][IMP] response to light stimulus [IMP] response to stress [IMP] |
| Molecular Function: |
succinate-semialdehyde dehydrogenase activity
[IDA][ISS]
3-chloroallyl aldehyde dehydrogenase activity [ISS] copper ion binding [IDA] NAD or NADH binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVIGAAARVA IGGCRKLISS HTSLLLVSSQ CRQMSMDAQS VSEKLRSSGL LRTQGLIGGK 60
61 WLDSYDNKTI KVNNPATGEI IADVACMGTK ETNDAIASSY EAFTSWSRLT AGERSKVLRR 120
121 WYDLLIAHKE ELGQLITLEQ GKPLKEAIGE VAYGASFIEY YAEEAKRVYG DIIPPNLSDR 180
181 RLLVLKQPVG VVGAITPWNF PLAMITRKVG PALASGCTVV VKPSELTPLT ALAAAELALQ 240
241 AGVPPGALNV VMGNAPEIGD ALLTSPQVRK ITFTGSTAVG KKLMAAAAPT VKKVSLELGG 300
301 NAPSIVFDDA DLDVAVKGTL AAKFRNSGQT CVCANRVLVQ DGIYDKFAEA FSEAVQKLEV 360
361 GDGFRDGTTQ GPLINDAAVQ KVETFVQDAV SKGAKIIIGG KRHSLGMTFY EPTVIRDVSD 420
421 NMIMSKEEIF GPVAPLIRFK TEEDAIRIAN DTIAGLAAYI FTNSVQRSWR VFEALEYGLV 480
481 GVNEGLISTE VAPFGGVKQS GLGREGSKYG MDEYLEIKYV CLGDMNRH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
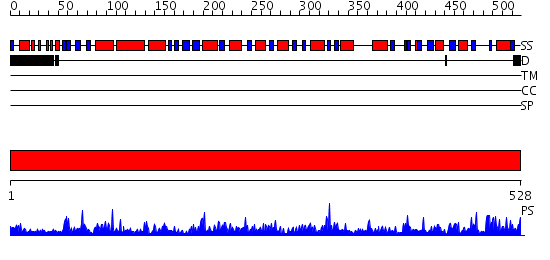
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..528] | 177.0 | Aldehyde reductase (dehydrogenase), ALDH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)