













| General Information: |
|
| Name(s) found: |
PLET2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
|
| Biological Process: |
root development
[IGI][IMP]
stem cell maintenance [IMP] regulation of transcription, DNA-dependent [ISS] pattern specification process [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSNNWLAFP LSPTHSSLPP HIHSSQNSHF NLGLVNDNID NPFQNQGWNM INPHGGGGEG 60
61 GEVPKVADFL GVSKSGDHHT DHNLVPYNDI HQTNASDYYF QTNSLLPTVV TCASNAPNNY 120
121 ELQESAHNLQ SLTLSMGSTG AAAAEVATVK ASPAETSADN SSSTTNTSGG AIVEATPRRT 180
181 LETFGQRTSI YRGVTRHRWT GRYEAHLWDN SCRREGQSRK GRQVYLGGYD KEEKAARAYD 240
241 LAALKYWGPS TTTNFPITNY EKEVEEMKNM TRQEFVASIR RKSSGFSRGA SMYRGVTRHH 300
301 QHGRWQARIG RVAGNKDLYL GTFSTEEEAA EAYDIAAIKF RGLNAVTNFE INRYDVKAIL 360
361 ESNTLPIGGG AAKRLKEAQA LESSRKREEM IALGSNFHQY GAASGSSSVA SSSRLQLQPY 420
421 PLSIQQPFEH LHHHQPLLTL QNNNDISQYH DSFSYIQTQL HLHQQQTNNY LQSSSHTSQL 480
481 YNAYLQSNPG LLHGFVSDNN NTSGFLGNNG IGIGSSSTVG SSAEEEFPAV KVDYDMPPSG 540
541 GATGYGGWNS GESAQGSNPG GVFTMWNE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
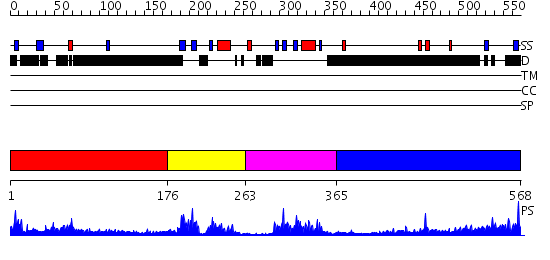
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [176..262] | 4.79 | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | |
| 3 | View Details | [263..364] | 22.0 | GCC-box binding domain | |
| 4 | View Details | [365..568] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.54 |
Source: Reynolds et al. (2008)