













| General Information: |
|
| Name(s) found: |
ROS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1393 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
DNA repair
[IMP]
DNA methylation [IDA] negative regulation of chromatin silencing [IMP] DNA demethylation [IMP] chromatin silencing [IMP] |
| Molecular Function: |
protein binding
[IPI]
DNA-(apurinic or apyrimidinic site) lyase activity [IDA][ISS] DNA N-glycosylase activity [IDA][ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKQRREESS FQQPPWIPQT PMKPFSPICP YTVEDQYHSS QLEERRFVGN KDMSGLDHLS 60
61 FGDLLALANT ASLIFSGQTP IPTRNTEVMQ KGTEEVESLS SVSNNVAEQI LKTPEKPKRK 120
121 KHRPKVRREA KPKREPKPRA PRKSVVTDGQ ESKTPKRKYV RKKVEVSKDQ DATPVESSAA 180
181 VETSTRPKRL CRRVLDFEAE NGENQTNGDI REAGEMESAL QEKQLDSGNQ ELKDCLLSAP 240
241 STPKRKRSQG KRKGVQPKKN GSNLEEVDIS MAQAAKRRQG PTCCDMNLSG IQYDEQCDYQ 300
301 KMHWLYSPNL QQGGMRYDAI CSKVFSGQQH NYVSAFHATC YSSTSQLSAN RVLTVEERRE 360
361 GIFQGRQESE LNVLSDKIDT PIKKKTTGHA RFRNLSSMNK LVEVPEHLTS GYCSKPQQNN 420
421 KILVDTRVTV SKKKPTKSEK SQTKQKNLLP NLCRFPPSFT GLSPDELWKR RNSIETISEL 480
481 LRLLDINREH SETALVPYTM NSQIVLFGGG AGAIVPVTPV KKPRPRPKVD LDDETDRVWK 540
541 LLLENINSEG VDGSDEQKAK WWEEERNVFR GRADSFIARM HLVQGDRRFT PWKGSVVDSV 600
601 VGVFLTQNVS DHLSSSAFMS LASQFPVPFV PSSNFDAGTS SMPSIQITYL DSEETMSSPP 660
661 DHNHSSVTLK NTQPDEEKDY VPSNETSRSS SEIAISAHES VDKTTDSKEY VDSDRKGSSV 720
721 EVDKTDEKCR VLNLFPSEDS ALTCQHSMVS DAPQNTERAG SSSEIDLEGE YRTSFMKLLQ 780
781 GVQVSLEDSN QVSPNMSPGD CSSEIKGFQS MKEPTKSSVD SSEPGCCSQQ DGDVLSCQKP 840
841 TLKEKGKKVL KEEKKAFDWD CLRREAQARA GIREKTRSTM DTVDWKAIRA ADVKEVAETI 900
901 KSRGMNHKLA ERIQGFLDRL VNDHGSIDLE WLRDVPPDKA KEYLLSFNGL GLKSVECVRL 960
961 LTLHHLAFPV DTNVGRIAVR LGWVPLQPLP ESLQLHLLEM YPMLESIQKY LWPRLCKLDQ1020
1021 KTLYELHYQM ITFGKVFCTK SKPNCNACPM KGECRHFASA FASARLALPS TEKGMGTPDK1080
1081 NPLPLHLPEP FQREQGSEVV QHSEPAKKVT CCEPIIEEPA SPEPETAEVS IADIEEAFFE1140
1141 DPEEIPTIRL NMDAFTSNLK KIMEHNKELQ DGNMSSALVA LTAETASLPM PKLKNISQLR1200
1201 TEHRVYELPD EHPLLAQLEK REPDDPCSYL LAIWTPGETA DSIQPSVSTC IFQANGMLCD1260
1261 EETCFSCNSI KETRSQIVRG TILIPCRTAM RGSFPLNGTY FQVNEVFADH ASSLNPINVP1320
1321 RELIWELPRR TVYFGTSVPT IFKGLSTEKI QACFWKGYVC VRGFDRKTRG PKPLIARLHF1380
1381 PASKLKGQQA NLA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
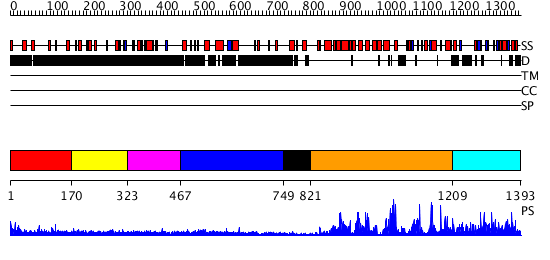
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 3.373996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [170..322] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [323..466] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [467..748] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [749..820] | 17.30103 | 8-oxoguanine glycosylase | |
| 6 | View Details | [821..1208] | 79.30103 | MutY adenine glycosylase in complex with DNA containing an A:oxoG pair | |
| 7 | View Details | [1209..1393] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)