













| General Information: |
|
| Name(s) found: |
gi|30725450
[NCBI NR]
gi|30696025 [NCBI NR] gi|22655274 [NCBI NR] gi|186531184 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 626 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPGIPLVSRE TSSCSRSTEQ MCHEDSRLRI SEEEEIAAEE SLAAYCKPVE LYNIIQRRAI 60
61 RNPLFLQRCL HYKIEAKHKR RIQMTVFLSG AIDAGVQTQK LFPLYILLAR LVSPKPVAEY 120
121 SAVYRFSRAC ILTGGLGVDG VSQAQANFLL PDMNRLALEA KSGSLAILFI SFAGAQNSQF 180
181 GIDSGKIHSG NIGGHCLWSK IPLQSLYASW QKSPNMDLGQ RVDTVSLVEM QPCFIKLKSM 240
241 SEEKCVSIQV PSNPLTSSSP QQVQVTISAE EVGSTEKSPY SSFSYNDISS SSLLQIIRLR 300
301 TGNVVFNYRY YNNKLQKTEV TEDFSCPFCL VKCASFKGLR YHLPSTHDLL NFEFWVTEEF 360
361 QAVNVSLKTE TMISKVNEDD VDPKQQTFFF SRRRQKSQVR SSRQGPHLGL GCEVLDKTDD 420
421 AHSVRSEKSR IPPGKHYERI GGAESGQRVP PGTSPADVQS CGDPDYVQSI AGSTMLQFAK 480
481 TRKISIERSD LRNRSLLQKR QFFHSHRAQP MALEQVLSDR DSEDEVDDDV ADFEDRRMLD 540
541 DFVDVTKDEK QMMHMWNSFV RKQRVLADGH IPWACEAFSR LHGPIMVRTP HLIWCWRVFM 600
601 VKLWNHGLLD ARTMNNCNTF LEQLQI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
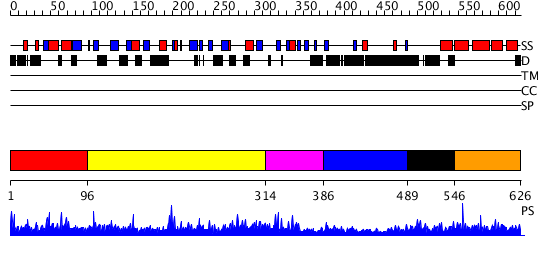
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 1.011977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [96..313] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [314..385] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [386..488] | 1.027993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [489..545] | 1.029997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [546..626] | 4.030994 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)