













| General Information: |
|
| Name(s) found: |
MAO2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] |
| Biological Process: |
malate metabolic process
[IEA][ISS]
metabolic process [IEA] oxidation reduction [IEA] |
| Molecular Function: |
ATP binding
[IDA]
zinc ion binding [IDA] malic enzyme activity [ISS] oxidoreductase activity, acting on NADH or NADPH, NAD or NADP as acceptor [ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMWKNIAGLS KAAAAARTHG SRRCFSTAIP GPCIVHKRGA DILHDPWFNK DTGFPLTERD 60
61 RLGIRGLLPP RVMTCVQQCD RFIESFRSLE NNTKGEPENV VALAKWRMLN RLHDRNETLY 120
121 YRVLIDNIKD FAPIIYTPTV GLVCQNYSGL YRRPRGMYFS AKDKGEMMSM IYNWPAPQVD 180
181 MIVITDGSRI LGLGDLGVQG IGIPIGKLDM YVAAAGINPQ RVLPIMLDVG TNNEKLLQND 240
241 LYLGVRQPRL EGEEYLEIID EFMEAAFTRW PKAVVQFEDF QAKWAFGTLE RYRKKFCMFN 300
301 DDVQGTAGVA LAGLLGTVRA QGRPISDFVN QKIVVVGAGS AGLGVTKMAV QAVARMAGIS 360
361 ESEATKNFYL IDKDGLVTTE RTKLDPGAVL FAKNPAEIRE GASIVEVVKK VRPHVLLGLS 420
421 GVGGIFNEEV LKAMRESDSC KPAIFAMSNP TLNAECTAAD AFKHAGGNIV FASGSPFENV 480
481 ELENGKVGHV NQANNMYLFP GIGLGTLLSG ARIVTDGMLQ AASECLASYM TDEEVQKGIL 540
541 YPSINNIRHI TAEVGAAVLR AAVTDDIAEG HGDVGPKDLS HMSKEDTVNY ITRNMWFPVY 600
601 SPLVHEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
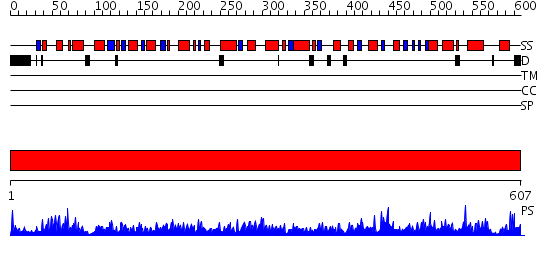
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..607] | 1000.0 | Crystal structure of a human malic enzyme |
| Source: Reynolds et al. 2008. Manuscript submitted |

|