













| General Information: |
|
| Name(s) found: |
IML1_YEAST
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1584 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFAKLHGKKQ RPISSINSQT PRTSNTTHAN SISLSSGNLI VGSNRNLRQK KEQFGSQQRA 60
61 SGRKLISNKE NDDNVNNGGD NNYDNGERVH RHHIPGLKIK AYQAELGYHE SRFSENLVML 120
121 NLVEFPDIKP GDLVELKTYH KNPSASNGDK KIYFIAKDFD GETKRRAKTS NVSILSGQLQ 180
181 TLLDLPSRSR IWIKLKPNKF DLQADVVEFN IKDCLLNRGD MWVLSSKLVD TCVFMDQRLA 240
241 FLDSIRGTIK GIYRNGKKIV SGYIGEQTRI IFRSESARLI FLIQITDEMW NFEETGEQLF 300
301 QKMVNSFFPK IFKKWKDVDT HHTITIAFAI SMDLSDTSFK DLTPGESLKN SQDYFRIVVD 360
361 QVSIIHWVDI METLREEFME IRKDLLNKQT DKGYSVANGR FSPVIKSNFL ELVNFATTIL 420
421 TDPFKQLDLR HTTTHVMIIS PGSGLFDVDY SLLRLTGKKL LSLEMTMDLI CLSKAPLHIV 480
481 PLFRYRDFEN KLHHCVPLWL SVFFWNDHDK KSNSEWTPRC KIYDLQMMGI TENELIREVD 540
541 VEYLQLNKKV KSLSEFMNDY DKNAFEVKIL CAGSNTKQSK KLNSKFDTVF ENDVVVKARK 600
601 IPATATTTHG NTKFIWRGPK VALPAIKDIQ KPNVIPDLSI KTIEASFYDD CNTTNDKIST 660
661 PTTSNNDNLE MNDSLVSVRS ADNQNTSLAL DSLKGLSKRN SLKDFTQRVI TKFISNIDTS 720
721 KNKKIKSTLL RDDVDNSPLG SNTPLPSSES KISGLKLQQK GLADENVISK RGNLIIKKNL 780
781 SIFGLPSNEI MSGSPSSYLG SSHTRTSSKL SNMSDKAAFI TEGQKSKHDD SNTYSLTQQL 840
841 KHRISETWVD IKSPSIPVSS EFANELLPIR WKDVWPKYVA RKYSKWRSFT TPAELPITIS 900
901 DFPSKDDFDR NFIFRNHSVT LNTDQEQYNQ TYKDLLRDMI YMRLLTGFQI CVGRQVEKIE 960
961 LSRESGESET VVNKYLDFNQ NDAFKLYLMI DSEIHRITCS SSGIIDVERY LRKDEANLFD1020
1021 QVPSYIPLVK TRYESSFRDA MIDPLHVKRE SLNWNQIDQV LAGYGDNLID RKWHGFRAKY1080
1081 VVLPTDIPPN TYSMVINGKS ETLNPEEIRV EGLRRLIGSI TRSRLRTEKE KKGRKTKREE1140
1141 IQPEVMFYTG PLYNFINEQQ TSLESSAINF KDSIFVNDNN LLNRNVELSK LAYQIQRGED1200
1201 RITLVNRKWH WKKHEKCFVG SEMVNWLIRN FSDIDTREDA IKYGQKVMKE GLFVHVLNKH1260
1261 NFLDGHYFYQ FSPEYVMDTN KLEKTNSHRS TLSDPKQMLR KASTGSSNDP SAMTPFSSVV1320
1321 PAISASNASV ADAKEPSRPI LMLSNSLVID VDPAGKSSKQ ESCTVHYDRV HNPDHCFHIR1380
1381 LEWLTTTPKL IDDLVGNWSR LCERYGLKMI EIPWEELCTI PSVNPFHSFV EIKLAINPWE1440
1441 DPEFKDRELF AKSKFYYHVY LLKASGFLLD NRASKFLQNQ DIEFDIMYSW GKPQFKYVQY1500
1501 IHHTGAYVAE LRENGCLFLA PNNIYISRVN PGNIIGKIHS ASSSSLDAQK VILNFKSTCL1560
1561 DYQKLRSIFL DAKEMWITGK IVED |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
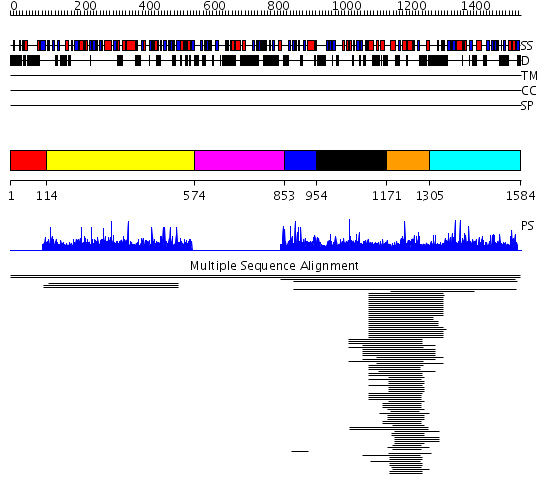
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [114..573] | 2.003826 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [574..852] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [853..953] | 1.005979 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [954..1170] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1171..1304] | 50.70927 | Segment polarity protein Dishevelled-1 | |
| 7 | View Details | [1305..1584] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [960..1151] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1152..1274] | 27.0 | Regulatory domain of epac2, domain 2; Regulatory domain of Epac2, domains 1 and 3 | |
| 10 | View Details | [1275..1337] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1338..1480] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1481..1584] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||
| 9 |
|
||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)