













| General Information: |
|
| Name(s) found: |
MAG2_YEAST
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVEPDMQKKA SGGSGGSEMD TLNATSNSSK QGVSNNKRNP VSKKKPGNKV SDGRDNAHNY 60
61 HGEGRRKSSK QQRSRTPYKE TSTRINDQDI DLSIQEEILG GNFKLRGRKT QVSINHLLNF 120
121 QLPEVEREKS RSSSSKKSNR RRDEHVHLHG DTFVNVNYRL LVDDRFDYPE QNCNPNVPVD 180
181 QEKILRVIVP KGQNCSICLS EEPVAPRMVT CGHIFCLSCL LNFFSIEETV KNKETGYSKK 240
241 KKYKECPLCG SIIGPKRVKP VLYEDDFDVT RLNQKPEPGA TVHLQLMCKP HGSLLPLPVA 300
301 LHLDPLKCGN FPPANLGSIK HYAHIMKCGV SYSLELYQKD IVAIQEQYEI DKAIYNDSGK 360
361 FVKQSIENIN DQISTLLAAT TDLSPLSNDI NNGLDNFHFD DDLLTKYDDS SAYFFYQTLV 420
421 ASSTKYFLSP LDVKILLTIF HYYSKFPESI ETTVENIHYD TVVTEQLIRR YKYIGHLPIG 480
481 TEIALLDLDW RKIPFLPKEI YEQFAHELKQ RRRKFTMKKQ KEDKEKKLYE KRLEQEHAEF 540
541 YRKENGNSLK FEDSVQMATH YESIVSSSIP LNSLGISMLG PPTNSCSTPQ KQAPSHTKRT 600
601 IWGTSIAVTE DEKASKENKE FQDMLLQRIR QEDSSDVTDS TDSPPTSNGK RGRKKKGKVM 660
661 LFSSNHQALG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
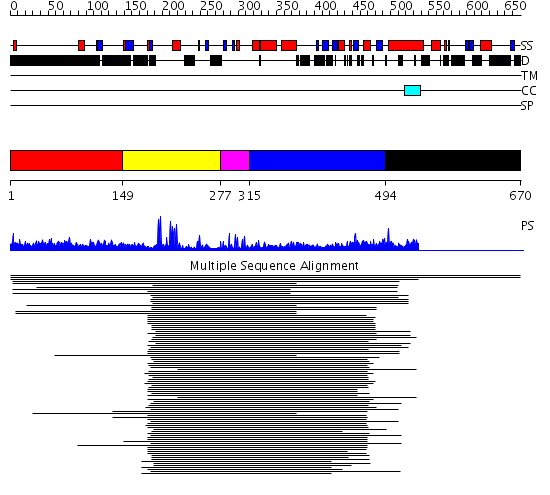
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..148] | 2.121992 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [149..276] | 4.221849 | bard1 RING domain | |
| 3 | View Details | [277..314] | 2.09691 | Nuclear factor XNF7 | |
| 4 | View Details | [315..493] | 5.279997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [494..670] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)