













| General Information: |
|
| Name(s) found: |
nuoB /
EG12083
[EcoGene]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 220 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane respiratory chain complex I
[IDA NADH dehydrogenase complex [IDA [NOT]cytoplasm [IDA membrane [IDA plasma membrane [IDA |
| Biological Process: |
anaerobic respiration
[IMP electron transport coupled proton transport [IMP photosynthesis, light reaction [IEA] oxidation reduction [IEA] aerobic respiration [IMP |
| Molecular Function: |
oxidoreduction-driven active transmembrane transporter activity
[IDA NADH dehydrogenase activity [IDA quinone binding [IEA] metal ion binding [IEA] iron ion binding [IEA] NADH dehydrogenase (ubiquinone) activity [IEA][IMP 4 iron, 4 sulfur cluster binding [IEA][IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYTLTRIDP NGENDRYPLQ KQEIVTDPLE QEVNKNVFMG KLNDMVNWGR KNSIWPYNFG 60
61 LSCCYVEMVT SFTAVHDVAR FGAEVLRASP RQADLMVVAG TCFTKMAPVI QRLYDQMLEP 120
121 KWVISMGACA NSGGMYDIYS VVQGVDKFIP VDVYIPGCPP RPEAYMQALM LLQESIGKER 180
181 RPLSWVVGDQ GVYRANMQSE RERKRGERIA VTNLRTPDEI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
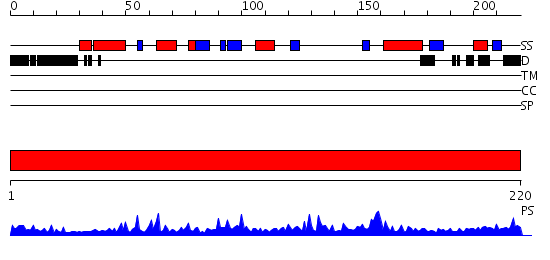
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | 53.522879 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)