













| General Information: |
|
| Name(s) found: |
murA /
EG11358
[EcoGene]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 419 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
cell division
[IEA]
regulation of cell shape [IEA] peptidoglycan biosynthetic process [IEA] cellular cell wall organization [IEA] UDP-N-acetylgalactosamine biosynthetic process [IEA] cell cycle [IEA] |
| Molecular Function: |
UDP-N-acetylglucosamine 1-carboxyvinyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKFRVQGPT KLQGEVTISG AKNAALPILF AALLAEEPVE IQNVPKLKDV DTSMKLLSQL 60
61 GAKVERNGSV HIDARDVNVF CAPYDLVKTM RASIWALGPL VARFGQGQVS LPGGCTIGAR 120
121 PVDLHISGLE QLGATIKLEE GYVKASVDGR LKGAHIVMDK VSVGATVTIM CAATLAEGTT 180
181 IIENAAREPE IVDTANFLIT LGAKISGQGT DRIVIEGVER LGGGVYRVLP DRIETGTFLV 240
241 AAAISRGKII CRNAQPDTLD AVLAKLRDAG ADIEVGEDWI SLDMHGKRPK AVNVRTAPHP 300
301 AFPTDMQAQF TLLNLVAEGT GFITETVFEN RFMHVPELSR MGAHAEIESN TVICHGVEKL 360
361 SGAQVMATDL RASASLVLAG CIAEGTTVVD RIYHIDRGYE RIEDKLRALG ANIERVKGE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
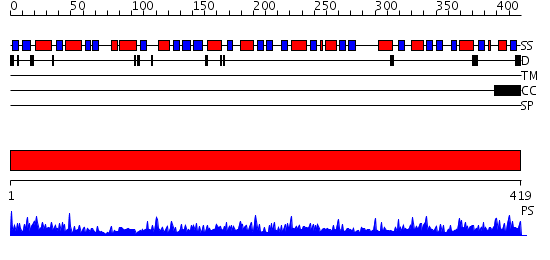
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..419] | 147.0 | UDP-N-acetylglucosamine enolpyruvyl transferase (EPT, MurA, MurZ) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)