













| General Information: |
|
| Name(s) found: |
P4KG6_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 622 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
inositol or phosphatidylinositol kinase activity
[ISS]
phosphotransferase activity, alcohol group as acceptor [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMAVFKAPL KGEFHGARKM EGKQYKHHLL QQQSTGRRRV FVQTDTGCVL GVELDRNDNV 60
61 HTVKKRLQIA FNFPTEESSL TFGDMVLKND LSAVRNDSPL LLKRNLMHRS SSTPCLSPTG 120
121 NDLQRKDRSG PIEILSHSPC FLSLKQTAND IVKAMKMGVE PIPVNGGLGG AYYFRDEKGQ 180
181 SVAIVKPTDE EPFAPNNPKG FVGKALGQPG LKPSVRVGET GFREVAAYLL DYDHFANVPP 240
241 TALVKITHSV FNVNDGMDGN KSREKKKLVS SKIASFQKFV PHDFDASDHG TSSFPVASVH 300
301 RIGILDIRIL NTDRHGGNLL VKKLDDGGVG RFGQVELIPI DHGLCLPETL EDPYFEWIHW 360
361 PQASIPFSEE ELDYIQSLDP VKDCEMLRRE LPMIREACLR VLVLCTVFLK EAAVFGLCLA 420
421 EIGEMMTREF RAGEEEPSEL EMLCIEAKRL TTEQDVLSPK SDGEGETEFQ FDIDYNELDS 480
481 VYGSETETDE FFAKNPFSNG RSSLGELKES IAEEEEDDEE EAKLTLSLSK LSTSMKNNLS 540
541 NTMGSGYLKP PKDNQTDKAL VSHKSANVQL PLSVNFVKLA DMKEVEWVVF LERFQELLYS 600
601 AFAERKTMTL RNTQRLGTSC KF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
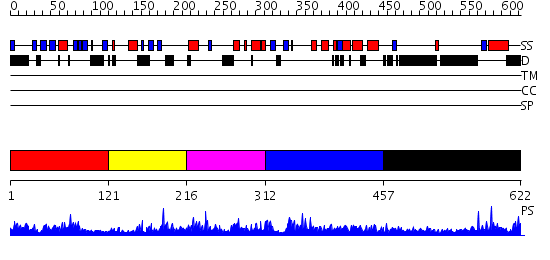
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 25.522879 | Solution Structure of S5a UIM-1/Ubiquitin Complex | |
| 2 | View Details | [121..215] | 2.07198 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [216..311] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [312..456] | 2.068961 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [457..622] | 1.030941 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)