













| General Information: |
|
| Name(s) found: |
gi|23506111
[NCBI NR]
gi|22329659 [NCBI NR] gi|17979007 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 700 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
heat shock protein binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKETPASSS MPSGLKAYTV PIFLFVLAMF FQLFLLPRSF PPSHYDVLGV KMYSSVDDVK 60
61 DAYQTVASKW DSGSGVSLPA DFVKIQYAYE LLTNLVWKRD YDLYAIDESV HIIEELEKQY 120
121 AVEDFAKIKL PLLEVVSYEP EREGFMSITS QDFASKFQDS KPWLIQVYSS GSNSSAQFAT 180
181 VWRRIVALLD GVANHAMLEL GDVQLVTYLA EKKPTGQVFF RKGLPSIFSF PPNCKTADCL 240
241 IRFEGELSAD AITDWFATTV LGLPRVFYHT KETLVSKFLS KVPPNKVKVI LFSQTGERAT 300
301 PIVRQAAKDY WNFASLSHVL WREEDASFWW NALEVESAPA IVIMKDPGSK PVVYHGSGNR 360
361 TWFLDILEQN KQLLLPQLRS TTSMELGCDA RGYSRAGYDK VTWYCAILVG RQSVELNKMR 420
421 ETMCRVQDAL SKYDDSDDAS KDPSITPAAS AHKSKRLSFA WLDGEAQNKY CFFYVQSETS 480
481 YDTCGTRRAP IDVPRILIVR YHRNATETAN AEQKSSKWPK TVWQSEADDV DPAAQLVVSY 540
541 DGTAEIPQII KWLSKMVDDG DNRNLPFYRA KTPELVPESA EPMRSGVPKT VKATQKLLSL 600
601 WNRIKDYLAD PRMGPTLLLG ALLSAGNVWW MRSRSSTQQP AQTNQPSPNQ PADNVEEKKK 660
661 KERKREQRRR NAKEEEVPAS ITDNEPKDAV QILSSGSDSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
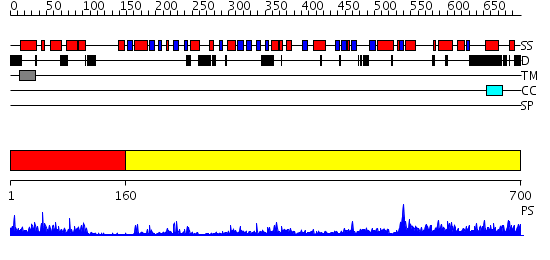
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 18.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [160..700] | 42.69897 | Crystal Structure of Yeast Protein Disulfide Isomerase |
| Source: Reynolds et al. 2008. Manuscript submitted |

|