













| General Information: |
|
| Name(s) found: |
gi|42572979
[NCBI NR]
gi|23505917 [NCBI NR] gi|16930419 [NCBI NR] gi|15234595 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 379 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule
[IDA]
|
| Biological Process: |
embryonic development
[IMP]
cell division [IMP] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPMESSSEQ DIVEESQKLV DSLDKLRVSA ASSSSNFKKK PIIIIVVGMA GSGKTSFLHR 60
61 LVCHTFDSKS HGYVVNLDPA VMSLPFGANI DIRDTVKYKE VMKQYNLGPN GGILTSLNLF 120
121 ATKFDEVVSV IEKRADQLDY VLVDTPGQIE IFTWSASGAI ITEAFASTFP TVVTYVVDTP 180
181 RSSSPITFMS NMLYACSILY KTRLPLVLAF NKTDVADHKF ALEWMEDFEV FQAAIQSDNS 240
241 YTATLANSLS LSLYEFYRNI RSVGVSAISG AGMDGFFKAI EASAEEYMET YKADLDMRKA 300
301 DKERLEEERK KHEMEKLRKD MESSQGGTVV LNTGLKDRDA TEKMMLEEDD EDFQVEDEED 360
361 SDDAIDEDDE DDETKHYYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
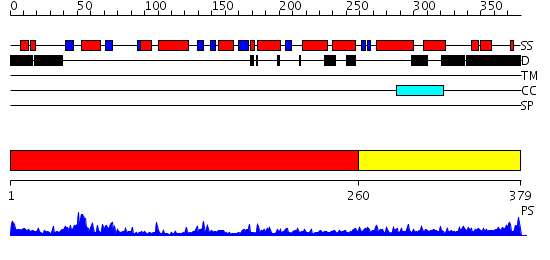
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | 27.30103 | PAB0955 crystal structure : a GTPase in Apo form from Pyrococcus abyssi | |
| 2 | View Details | [260..379] | 20.39794 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)