













| General Information: |
|
| Name(s) found: |
APC2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 865 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[TAS]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
ubiquitin protein ligase binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEALGSSDCN LEILETLSDD AIQEITESYD GFFTTVESLI AGTGDSLVED EFVSHVYCLC 60
61 KYGLDSLVRD HFLRSLEQAF EKGGASSFWQ HFDAYSEKKH HNYGEEIQIV LCKALEEISI 120
121 EKQYHEKCLS IVVHALQSFK EQSSDDRQNS DTERVHLFSR FQSMLSSTLM TTLPQHFPEI 180
181 LHWYFKERLE ELSAIMDGDG IEEQEDDCMD LDEKLRYKNG EMDVDEGCSQ GKRLGHDKLV 240
241 KNIGKVVRDL RSIGFTSMAE NAYASAIFLL LKAKVHDLAG DDYRTSVLES IKEWIQTVPL 300
301 QFLNALLSYL GDSVSYGTTS SGLTSPLACC PSPSFSRVVT PSEGIVRWKL RLEYFAYETL 360
361 QDLRIAKLFE IIVDYPESSP AIEDLKQCLE YTGQHSKLVE SFISSLKYRL LTAGASTNDI 420
421 LHQYVSTIKA LRAIDPAGVF LEAVGEPIRD YLRGRKDTIK CIVTMLTDGS GGNANGSGNP 480
481 GDSLLEELMR DEESQENVGF DDDFHTDDKQ AWINASRWEP DPVEADPLKG SLSQRKVDIL 540
541 GMLVDIIGSK EQLVNEYRVM LAEKLLNKTD YDIDTEIRTV ELLKIHFGEA SMQRCEIMLN 600
601 DLIDSKRVNT NIKKASQTGA ELRENELSVD TLTSTILSTN FWPPIQDEPL ELPGPVDKLL 660
661 SDYANRYHEI KTPRKLLWKK NLGTVKLELQ FEDRAMQFTV SPTHAAIIMQ FQEKKSWTYK 720
721 DLAEVIGIPI DALNRRVNFW ISKGVLREST GANSNSSVLT LVESITDSGK NEGEELLTGE 780
781 EEGETSIASV EDQLRKEMTI YEKFIMGMLT NFGSMALERI HNTLKMFCVA DPSYDKSLQQ 840
841 LQSFLSGLVS EEKLEFRDGM YLLKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
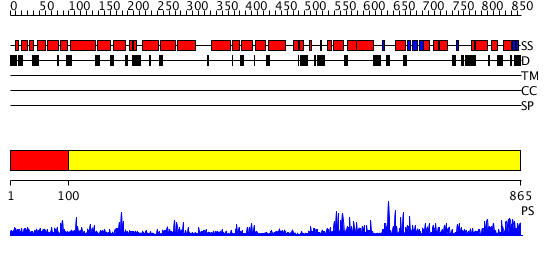
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..865] | 152.0 | Crystal Structure of The Cand1-Cul1-Roc1 Complex |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)