













| General Information: |
|
| Name(s) found: |
GGAP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 442 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
callose deposition in cell wall during defense response
[IMP]
response to heat [IMP] response to jasmonic acid stimulus [IEP] L-ascorbic acid biosynthetic process [IMP] defense response to bacterium [IMP] |
| Molecular Function: |
GDP-galactose:mannose-1-phosphate guanyltransferase activity
[IDA]
GDP-galactose:myoinositol-1-phosphate guanyltransferase activity [IDA] GDP-galactose:glucose-1-phosphate guanyltransferase activity [IDA] mannose-1-phosphate guanylyltransferase (GDP) activity [IDA] quercetin 4'-O-glucosyltransferase activity [IDA] GDP-D-glucose phosphorylase activity [IDA] galactose-1-phosphate guanylyltransferase (GDP) activity [IDA] glucose-1-phosphate guanylyltransferase (GDP) activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKIKRVPTV VSNYQKDDGA EDPVGCGRNC LGACCLNGAR LPLYACKNLV KSGEKLVISH 60
61 EAIEPPVAFL ESLVLGEWED RFQRGLFRYD VTACETKVIP GKYGFVAQLN EGRHLKKRPT 120
121 EFRVDKVLQS FDGSKFNFTK VGQEELLFQF EAGEDAQVQF FPCMPIDPEN SPSVVAINVS 180
181 PIEYGHVLLI PRVLDCLPQR IDHKSLLLAV HMAAEAANPY FRLGYNSLGA FATINHLHFQ 240
241 AYYLAMPFPL EKAPTKKITT TVSGVKISEL LSYPVRSLLF EGGSSMQELS DTVSDCCVCL 300
301 QNNNIPFNIL ISDCGRQIFL MPQCYAEKQA LGEVSPEVLE TQVNPAVWEI SGHMVLKRKE 360
361 DYEGASEDNA WRLLAEASLS EERFKEVTAL AFEAIGCSNQ EEDLEGTIVH QQNSSGNVNQ 420
421 KSNRTHGGPI TNGTAAECLV LQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
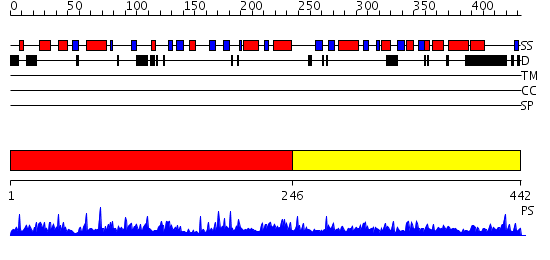
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 1.2 | X-ray structure of galt-like protein from arabidopsis thaliana at5g18200 | |
| 2 | View Details | [246..442] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)