













| General Information: |
|
| Name(s) found: |
AMY3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 887 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] |
| Biological Process: |
starch catabolic process
[TAS]
|
| Molecular Function: |
alpha-amylase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTVPIESLL HHSYLRHNSK VNRGNRSFIP ISLNLRSHFT SNKLLHSIGK SVGVSSMNKS 60
61 PVAIRATSSD TAVVETAQSD DVIFKEIFPV QRIEKAEGKI YVRLKEVKEK NWELSVGCSI 120
121 PGKWILHWGV SYVGDTGSEW DQPPEDMRPP GSIAIKDYAI ETPLKKLSEG DSFFEVAINL 180
181 NLESSVAALN FVLKDEETGA WYQHKGRDFK VPLVDDVPDN GNLIGAKKGF GALGQLSNIP 240
241 LKQDKSSAET DSIEERKGLQ EFYEEMPISK RVADDNSVSV TARKCPETSK NIVSIETDLP 300
301 GDVTVHWGVC KNGTKKWEIP SEPYPEETSL FKNKALRTRL QRKDDGNGSF GLFSLDGKLE 360
361 GLCFVLKLNE NTWLNYRGED FYVPFLTSSS SPVETEAAQV SKPKRKTDKE VSASGFTKEI 420
421 ITEIRNLAID ISSHKNQKTN VKEVQENILQ EIEKLAAEAY SIFRSTTPAF SEEGVLEAEA 480
481 DKPDIKISSG TGSGFEILCQ GFNWESNKSG RWYLELQEKA DELASLGFTV LWLPPPTESV 540
541 SPEGYMPKDL YNLNSRYGTI DELKDTVKKF HKVGIKVLGD AVLNHRCAHF KNQNGVWNLF 600
601 GGRLNWDDRA VVADDPHFQG RGNKSSGDNF HAAPNIDHSQ DFVRKDIKEW LCWMMEEVGY 660
661 DGWRLDFVRG FWGGYVKDYM DASKPYFAVG EYWDSLSYTY GEMDYNQDAH RQRIVDWINA 720
721 TSGAAGAFDV TTKGILHTAL QKCEYWRLSD PKGKPPGVVG WWPSRAVTFI ENHDTGSTQG 780
781 HWRFPEGKEM QGYAYILTHP GTPAVFFDHI FSDYHSEIAA LLSLRNRQKL HCRSEVNIDK 840
841 SERDVYAAII DEKVAMKIGP GHYEPPNGSQ NWSVAVEGRD YKVWETS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
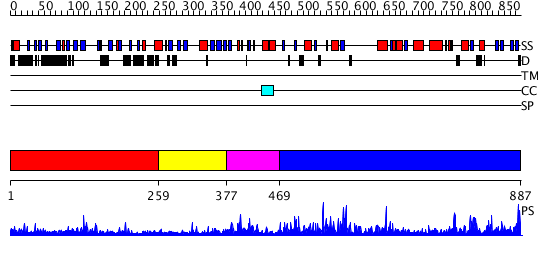
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 17.0 | No description for 2j44A was found. | |
| 2 | View Details | [259..376] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [377..468] | 1.567995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [469..887] | 54.0 | CRYSTAL AND MOLECULAR STRUCTURE OF BARLEY ALPHA-AMYLASE |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)