













| General Information: |
|
| Name(s) found: |
gi|42567721
[NCBI NR]
gi|30102672 [NCBI NR] gi|110743660 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1084 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA repair
[IEA]
|
| Molecular Function: |
nucleic acid binding
[IEA]
zinc ion binding [IEA] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] phosphoric diester hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRRQCEKVV IRIHNIGTPL ISGSSGLPLE LFHIQSDRPY TIGRSSSDGF CDFVIDHSSI 60
61 SRKHCQILFD SQSHKLYIFD GVIHLPSGSF SQVYDEFRRR LVGVEDLGNL KFRASLNGVY 120
121 VNRVRVRKSK VQEVSIDDEV LFFCGKEGLC CKDGRVGFVV QEIVFEGRDA SIVSVSSGHS 180
181 RGTFSSGKRS KRVFAPMENE INSPVSGFYP PKAVGVVERV NSLVSYCRHI LKSDDPLSCL 240
241 RLSIISHSGK ECLSCCTSKM FRSKVGIVAD DRGVKSAEIN HDMGHGLSGL RLSIERPNSN 300
301 LHVDRRLGVS DLISEIENEF AACTFISDKT RTMLPFDGEK VNTPDITCIN KEKSYQSSLQ 360
361 APGKNFYLNR LQYIEQSSTG CQRVVSLPEL LHPVESIQQI FLATFTSDIL WFLTCCDTPR 420
421 HLPVTIACHN AERCWSSNPD ARTAVPLPNY PNVTMVYPPF PEEIAFGKDR TNRGIACHHP 480
481 KLFILQRKDS IRVIITSANL VARQWNDVTN TVWWQDFPRR ADPDLLSLFG HCQRETNHGL 540
541 KPDFCAQLAG FAASLLTDVP SQAHWILEFT KYNFEHSAGH LVASVPGIHS YKPSYLTESG 600
601 CSNTIFSEEF LGSVEASVVG LSYLFRSAND STGAQLKRLA SYIRRTRENS LGMLELVLRR 660
661 NTNVPADPNA VRVLVPNPDD DSRDDFVQLG FLPRSIAKWV SPLWDIGFFK FVGYVYRDEV 720
721 LGAASCRSNE KVQLVLHVLQ GVSISDMSKL IQPYHVVALC SLIASLQRCT GIWRLQEVLG 780
781 RYKWPESQES DFVYSASSIG GSATTGFQAD FSSAAGKKAL QHFDSQESDP EWGCWSNREE 840
841 REAPSIKIIF PTIERVKNGH HGVLSSRRLL CFSEKTWQKW RHSNVLHDAV PNPQDRVGHP 900
901 MHIKVARRLF TSTRGSRSSS FGWVYSGSHN FSAAAWGQTI SRSSRNNQDQ SNNAIRAVKK 960
961 LRVCNYELGI VFVFPPPHEE TDSCEGSKID DIVLPFVVPA PKYGWSDKPA TGLAMREAFA1020
1021 EFREGSTSFC GESEVEEEVE EEEEEEADAE GRGEFVAEEE KQEEEAYAEA LWSQVESSSS1080
1081 SLSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
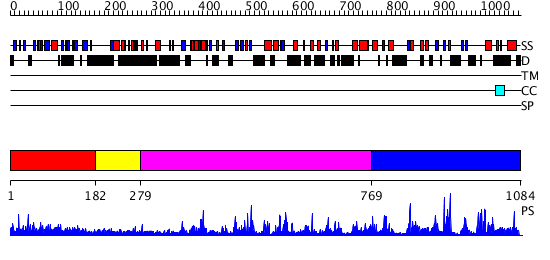
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 13.79588 | No description for PF00498.18 was found. No confident structure predictions are available. | |
| 2 | View Details | [182..278] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [279..768] | 50.69897 | Crystal Structure Analysis of the Yeast Tyrosyl-DNA Phosphodiesterase | |
| 4 | View Details | [769..1084] | 102.0 | human Tyrosyl DNA phosphodiesterase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.44 |
Source: Reynolds et al. (2008)