













| General Information: |
|
| Name(s) found: |
CAAT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[ISS]
plasma membrane [IDA] |
| Biological Process: |
L-glutamate import
[IGI]
basic amino acid transport [IGI] L-arginine import [IGI] |
| Molecular Function: |
L-lysine transmembrane transporter activity
[IGI]
arginine transmembrane transporter activity [IGI] cationic amino acid transmembrane transporter activity [IGI][ISS] L-glutamate transmembrane transporter activity [IGI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASGGGDDGL RRRGCSCTKD DFLPEESFQS MGNYLKALKE TPSRFMDRIM TRSLDSDEIN 60
61 EMKARSGHEM KKTLTWWDLM WFGIGAVIGS GIFVLTGLEA RNHSGPAVVL SYVVSGVSAM 120
121 LSVFCYTEFA VEIPVAGGSF AYLRVELGDF MAFIAAGNII LEYVVGGAAV ARSWTSYFAT 180
181 LLNHKPEDFR IIVHKLGEDY SHLDPIAVGV CAIICVLAVV GTKGSSRFNY IASIIHMVVI 240
241 LFVIIAGFTK ADVKNYSDFT PYGVRGVFKS AAVLFFAYIG FDAVSTMAEE TKNPGRDIPI 300
301 GLVGSMVVTT VCYCLMAVTL CLMQPYQQID PDAPFSVAFS AVGWDWAKYI VAFGALKGMT 360
361 TVLLVGAIGQ ARYMTHIARA HMMPPWLAQV NAKTGTPINA TVVMLAATAL IAFFTKLKIL 420
421 ADLLSVSTLF IFMFVAVALL VRRYYVTGET STRDRNKFLV FLGLILASST ATAVYWALEE 480
481 EGWIGYCITV PIWFLSTVAM KFLVPQARAP KIWGVPLVPW LPSASIAINI FLLGSIDTKS 540
541 FVRFAIWTGI LLIYYVLFGL HATYDTAKAT LKEKQALQKA EEGGVVADNS CSAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
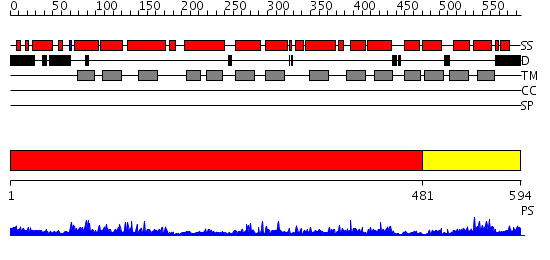
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..480] | 1.36 | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | |
| 2 | View Details | [481..594] | 2.096992 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|