













| General Information: |
|
| Name(s) found: |
ATG7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 697 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
autophagy
[ISS]
leaf senescence [TAS] protein amino acid lipidation [ISS] senescence [IMP] |
| Molecular Function: |
APG8 activating enzyme activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEKETPAII LQFAPLNSSV DEGFWHSFSS LKLDKLGIDD SPISITGFYG PCGHPQVSNH 60
61 LTLLSESLPL DEQSLIASTS HGNRNKCPVP GILYNTNTVE SFNKLDKQSL LKAEANKIWE 120
121 DIQSGKALED PSVLPRFLVI SFADLKKWSF RYWFAFPAFV LDPPVSLIEL KPASEYFSSE 180
181 EAESVSAACN DWRDSDLTTD VPFFLVSVSS DSKASIRHLK DLEACQGDHQ KLLFGFYDPC 240
241 HLPSNPGWPL RNYLALIRSR WNLETVWFFC YRESRGFADL NLSLVGQASI TLSSGESAET 300
301 VPNSVGWELN KGKRVPRSIS LANSMDPTRL AVSAVDLNLK LMRWRALPSL NLNVLSSVKC 360
361 LLLGAGTLGC QVARTLMGWG IRNITFVDYG KVAMSNPVRQ SLYNFEDCLG RGEFKAVAAV 420
421 KSLKQIFPAM ETSGVVMAIP MPGHPISSQE EDSVLGDCKR LSELIESHDA VFLLTDTRES 480
481 RWLPSLLCAN ANKIAINAAL GFDSYMVMRH GAGPTSLSDD MQNLDINKTN TQRLGCYFCN 540
541 DVVAPQDSMT DRTLDQQCTV TRPGLAPIAG ALAVELLVGV LQHPLGINAK GDNSSLSNTG 600
601 NNDDSPLGIL PHQIRGSVSQ FSQITLLGQA SNSCTACSET VISEYRERGN SFILEAINHP 660
661 TYLEDLTGLT ELKKAANSFN LDWEDDDTDD DDVAVDL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
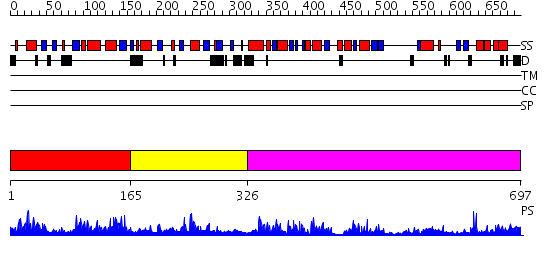
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 2.046977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [165..325] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [326..697] | 54.69897 | Molybdenum cofactor biosynthesis protein MoeB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)