













| General Information: |
|
| Name(s) found: |
gi|21553726
[NCBI NR]
gi|18413696 [NCBI NR] gi|110738711 [NCBI NR] |
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] plasma membrane [IDA] |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATASSSESP SEETQTNQEA MQQQDLPKET TTEEEILYKT KSIQFLGRTT PIILQNENGP 60
61 CPLLAICNVL LLRNNLNLNP DCYEVSQERL MSLVVDRLID SNSKVNNKDE GYIENQQQNI 120
121 ADAIDLLPRL TTGIDVNIKF RRIDDFEFTP ECAIFDLLDI PLYHGWIVDP QDVEAANAIG 180
181 SKSYNALMGE LVALETQNVE AQGDQNPGED SVDFAAATTA VLGVPSPCLS KTRSFDDSPP 240
241 AAAELRRMRK GDLEEETELL QALQLSQGQG NDPTPNTNGD STNQDSAFTF SDASPTSTHI 300
301 TNISQLDQFK TDDDKASEND GNMIKVGKFP TTITTKSEDL NHDQLSSKQS GGETACDVEN 360
361 VSSSKEAIVD VTSSEALSVD KGNLESAKSE SSSESVLKSD AASIDPDLSC RSQHDDAPNA 420
421 FIPPVSTDEP MYEGEECVNT VPPVCADKEP VYEGESLLGK RVEKDVGDCS SEGRATDGLT 480
481 AEEGELIRNF MKNSASQLTF CGLFRLQEGL KERELCVFFR NNHFCTMFKY EGELYLLATD 540
541 QGYLNQPDLV WEKLNEVNGD TAFMTATFKD FTIDSSTGGA SGTWDERNAV TNTADYLASI 600
601 NNVADAGIDV NSDLQLAIAL QQQEFEDQSP RSNPTPQPTS VAASRLVTGP QVPRSSHRPS 660
661 SAASSRHDGK SPKDKDSKCR IM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
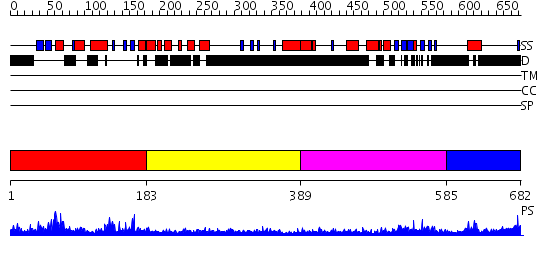
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 75.677781 | No description for PF04424.4 was found. No confident structure predictions are available. | |
| 2 | View Details | [183..388] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [389..584] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [585..682] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)