













| General Information: |
|
| Name(s) found: |
SPA4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 794 amino acids |
Gene Ontology: |
|
| Cellular Component: |
heterotrimeric G-protein complex
[ISS]
CUL4 RING ubiquitin ligase complex [ISS] |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
protein binding
[IPI]
signal transducer activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGSSESSSR GLNNTSGVSE FCTDGSKSLS HIDYVRSLLG SHKEANLGGL DDDSIVRALE 60
61 CEDVSLRQWL DNPDRSVDAF ECFHVFRQIV EIVNAAHSQG IVVHNVRPSC FVMSSFNNVS 120
121 FIESASCSDS GSDEDATTKS REIGSSRQEE ILSERRSKQQ EEVKKQPFPM KQILAMEMSW 180
181 YTSHEEDNGS LCNCASDIYR LGVLLFELFC PVSSREEKSR TMSSLRHRVL PPQILLNWPK 240
241 EASFCLWLLH PEPSCRPSMS ELLQSEFINE PRENLEEREA AMELRDRIEE QELLLEFLFL 300
301 IQQRKQEAAD KLQDTISLLS SDIDQVVKRQ LVLQQKGRDV RSFLASRKRI RQGAETTAAE 360
361 EENDDNSIDE ESKLDDTLES TLLESSRLMR NLKKLESVYF ATRYRQIKAA TAAEKPLARY 420
421 YSALSCNGRS SEKSSMSQPS KDPINDSRQG GWIDPFLEGL CKYLSFSKLR VKADLKQGDL 480
481 LNSSNLVCAI GFDRDGEFFA TAGVNKKIKI FECESIIKDG RDIHYPVVEL ASRSKLSGIC 540
541 WNSYIKSQVA SSNFEGVVQV WDVARNQLVT EMKEHEKRVW SIDYSSADPT LLASGSDDGS 600
601 VKLWSINQGV SIGTIKTKAN ICCVQFPSET GRSLAFGSAD HKVYYYDLRN PKLPLCTMIG 660
661 HHKTVSYVRF VDSSTLVSSS TDNTLKLWDL SMSISGINET PLHSFMGHTN VKNFVGLSVS 720
721 DGYIATGSET NEVFVYHKAF PMPVLSYKFK TIDPVSELEV DDASQFISSV CWRGQSSTLV 780
781 AANSTGNIKI LEMV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
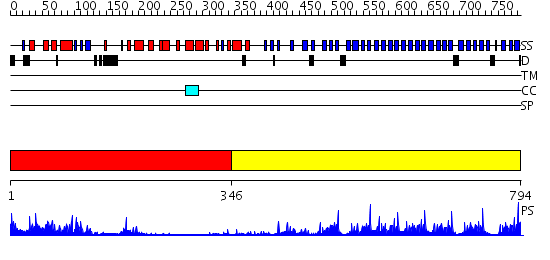
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..345] | 28.0 | Crystal structure of non-phosphorylated Fus3 | |
| 2 | View Details | [346..794] | 59.69897 | F-box/WD-repeat protein 1 (beta-TRCP1) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)