













| General Information: |
|
| Name(s) found: |
ESP3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1044 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
RNA splicing
[NAS]
posttranscriptional gene silencing by RNA [IMP] embryonic development ending in seed dormancy [NAS] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA] ATP-dependent RNA helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASNDLKTWV SDKLMMLLGY SQAAVVNYLI AMAKKTKSPT ELVGELVDYG FSSSGDTRSF 60
61 AEEIFARVPR KTAGVNLYQK HEAEAAMLVR KQKTYALLDA DDDEDEVVVE KKSSVSESRK 120
121 SDKGKKRFRK KSGQSDESDG EVAVREDSRH VRRKVSEEDD GSESEEERVR DQKEREELEQ 180
181 HLKDRDTART RKLTEQTLSK KEKEEAVRRA NALEKDDLYS LRKVSRQEYL KKREQKKLDE 240
241 LRDEIEDEQY LFGGEKLTET ELREFRYKKE LYDLVKKRTQ DEDNVEEYRI PDAYDQEGGV 300
301 DQEKRFSVAV QRYRDLDSTE KMNPFAEQEA WEDHQIGKAT LKFGAKNKQA SDDYQFVFED 360
361 QINFIKESVM AGENYEDAMD AKQKSQDLAE KTALEELQEV RRSLPIYTYR DQLLKAVEEH 420
421 QVLVIVGDTG SGKTTQIPQY LHEAGYTKRG KVGCTQPRRV AAMSVAARVA QEMGVKLGHE 480
481 VGYSIRFEDC TSDKTVLKYM TDGMLLRELL GEPDLASYSV VIVDEAHERT LSTDILFGLV 540
541 KDIARFRPDL KLLISSATMD AEKFSDYFDT APIFSFPGRR YPVEINYTSA PEADYMDAAI 600
601 VTILTIHVRE PLGDILVFFT GQEEIETAEE ILKHRIRGLG TKIRELIICP IYANLPSELQ 660
661 AKIFEPTPEG ARKVVLATNI AETSLTIDGI KYVVDPGFSK MKSYNPRTGM ESLLITPISK 720
721 ASATQRAGRA GRTSPGKCYR LYTAFNYNND LEENTVPEVQ RTNLASVVLA LKSLGIHDLI 780
781 NFDFMDPPPA EALVKSLELL FALGALNKLG ELTKAGRRMA EFPLDPMLSK MIVVSDKYKC 840
841 SDEIISIAAM LSIGGSIFYR PKDKQVHADN ARMNFHTGNV GDHIALLKVY SSWKETNFST 900
901 QWCYENYIQV RSMKRARDIR DQLEGLLERV EIDISSNLNE LDSVRKSIVA GFFPHTAKLQ 960
961 KNGSYRTVKH PQTVHIHPNS GLSQVLPRWV VYHELVLTSK EYMRQVTELK PEWLIELAPH1020
1021 YYQLKDVEDA ASKKMPKGAG KAAM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
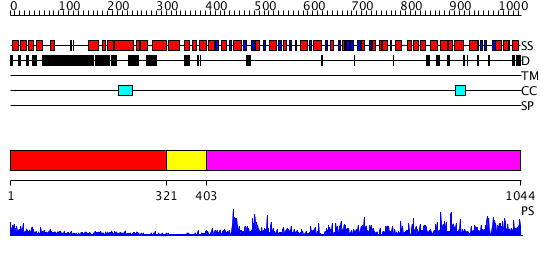
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..320] | 6.39794 | Heavy meromyosin subfragment | |
| 2 | View Details | [321..402] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [403..1044] | 20.09691 | No description for 2p6rA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)