













| General Information: |
|
| Name(s) found: |
LPAH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMTEATKVLY IVVREEGDDD DNNGDDSFRY TRPVLQSTLQ LMGCKARHAF KISRRVFELI 60
61 RSEGSCNTSP ENGKEPEFAK EVGGSTCVEK LNCLVVAGDV DKNKSKPFEM YKRRTTVVVS 120
121 REIFVDVVCD ALAEYKYVGR DQRADLILAC RIRERKESVT VLLCGTSGCG KSTLSALLGS 180
181 RLGITTVVST DSIRHMMRSF ADEKQNPLLW ASTYHAGEYL DPVAVAESKA KRKAKKLKGS 240
241 RGVNSNAQKT DAGSNSSTTE LLSHKQMAIE GYKAQSEMVI DSLDRLITTW EERNESVVVE 300
301 GVHLSLNFVM GLMKKHPSIV PFMVYIANEE KHLERFAVRA KYMTLDPAKN KYVKYIRNIR 360
361 TIQDYLCKRA DKHLVPKINN TNVDKSVATI HATVFGCLRR RETGEKLYDT TTNTVSVIDD 420
421 EHRNQCAANS LTSKGMFQVI QRQGSSRRFM ALCNTDGTVA KTWPVASVGK IRKPVVNTEM 480
481 DDGTEHQLHK AEPVNLQFGH FGISAWPSDG ATSHAGSVDD LRADIIETGS RHYSSCCSSP 540
541 RTSDGPSKEL MEEQSVNGSD EDDEEGDDDF HEPDSDEDLS DNNDERNRDE IGSVDEESTK 600
601 SDEEYDDLAM EDKSYWTDNE EEESRDTISM VSQNNHNEAS KTNKDDKYSQ NLDLFLKTTN 660
661 QPLTESLELT SEYRNRMGVA ASDKAKMRKR SLSIPPVGKH GSIIDDQILA NQTDSVL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
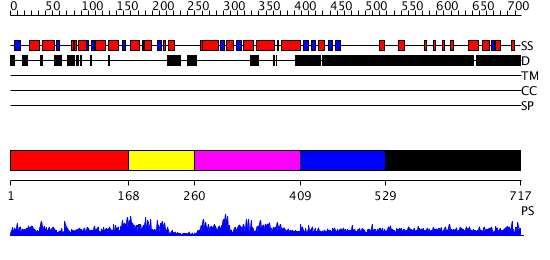
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 3.39794 | Signal sequence recognition protein Ffh; GTPase domain of the signal sequence recognition protein Ffh | |
| 2 | View Details | [168..259] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [260..408] | 2.051979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [409..528] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [529..717] | 5.100997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)