













| General Information: |
|
| Name(s) found: |
CHLD_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
magnesium chelatase complex [TAS] |
| Biological Process: |
chlorophyll biosynthetic process
[TAS]
|
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] nucleoside-triphosphatase activity [IEA] magnesium chelatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMTPVASSS PVSTCRLFRC NLLPDLLPKP LFLSLPKRNR IASCRFTVRA SANATVESPN 60
61 GVPASTSDTD TETDTTSYGR QFFPLAAVVG QEGIKTALLL GAVDREIGGI AISGRRGTAK 120
121 TVMARGLHEI LPPIEVVVGS ISNADPACPD EWEDDLDERI EYNADNTIKT EIVKSPFIQI 180
181 PLGVTEDRLI GSVDVEESVK RGTTVFQPGL LAEAHRGVLY VDEINLLDEG ISNLLLNVLT 240
241 DGVNIVEREG ISFRHPCKPL LIATYNPEEG AVREHLLDRV AINLSADLPM SFEDRVAAVG 300
301 IATQFQERCN EVFRMVNEET ETAKTQIILA REYLKDVKIS REQLKYLVLE AVRGGVQGHR 360
361 AELYAARVAK CLAAIEGREK VTIDDLRKAV ELVILPRSSL DETPPEQQNQ PPPPPPPPQN 420
421 SESGEEENEE EQEEEEEDES NEENENEQQQ DQIPEEFIFD AEGGLVDEKL LFFAQQAQKR 480
481 RGKAGRAKNV IFSEDRGRYI KPMLPKGPVK RLAVDATLRA AAPYQKLRRE KDISGTRKVF 540
541 VEKTDMRAKR MARKAGALVI FVVDASGSMA LNRMQNAKGA ALKLLAESYT SRDQVSIIPF 600
601 RGDAAEVLLP PSRSIAMARN RLERLPCGGG SPLAHGLTTA VRVGLNAEKS GDVGRIMIVA 660
661 ITDGRANITL KRSTDPESIA PDAPRPTSKE LKDEILEVAG KIYKAGMSLL VIDTENKFVS 720
721 TGFAKEIARV AQGKYYYLPN ASDAVISATT RDALSDLKNS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
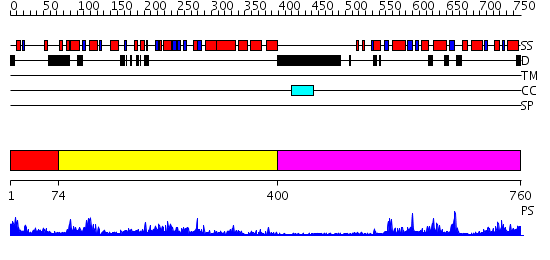
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 8.0 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 2 | View Details | [74..399] | 51.69897 | ATPase subunit of magnesium chelatase, BchI | |
| 3 | View Details | [400..760] | 14.0 | No description for 2ok5A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)