













| General Information: |
|
| Name(s) found: |
HAT31_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 723 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
positive regulation of transcription [IDA] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYKAVSKRVT RSSGSGLKQT NVDNGGEISP TVDRVSEQGK SSEAGSHMPT DANGNGHLHH 60
61 EIMDHGKGNE EQKPTPQTVK KDSNTNTKFS GSHRELVIGL PCRGQFEIHN RSRASTSSKR 120
121 LGGGGERNVL FASHKRAQRS KEDAGPSSVV ANSTPVGRPK KKNKTMNKGQ VREDDEYTRI 180
181 KKKLRYFLNR INYEQSLIDA YSLEGWKGSS LEKIRPEKEL ERATKEILRR KLKIRDLFQH 240
241 LDTLCAEGSL PESLFDTDGE ISSEDIFCAK CGSKDLSVDN DIILCDGFCD RGFHQYCLEP 300
301 PLRKEDIPPD DEGWLCPGCD CKDDSLDLLN DSLGTKFSVS DSWEKIFPEA AAALVGGGQN 360
361 LDCDLPSDDS DDEEYDPDCL NDNENDEDGS DDNEESENED GSSDETEFTS ASDEMIESFK 420
421 EGKDIMKDVM ALPSDDSEDD DYDPDAPTCD DDKESSNSDC TSDTEDLETS FKGDETNQQA 480
481 EDTPLEDPGR QTSQLQGDAI LESDVGLDDG PAGVSRRRNV ERLDYKKLYD EEYDNVPTSS 540
541 SDDDDWDKTA RMGKEDSESE DEGDTVPLKQ SSNAEDHTSK KLIRKSKRAD KKDTLEMPQE 600
601 GPGENGGSGE IEKSSSSACK QTDPKTQRLY ISFQENQYPD KATKESLAKE LQMTVKQVNN 660
661 WFKHRRWSIN SKPLVSEENV EKLKTGKEGE CETSVAGSSK QTMETESVAE KPTNTGSRKR 720
721 RRK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
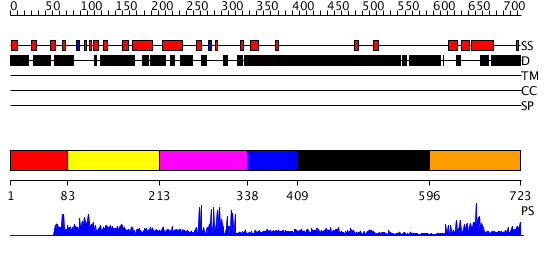
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..212] | 1.100974 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [213..337] | 10.39794 | No description for 2ysmA was found. | |
| 4 | View Details | [338..408] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [409..595] | 2.930997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [596..723] | 20.522879 | Ultrabithorax (ubx) homeodomain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)