













| General Information: |
|
| Name(s) found: |
gi|241908527
[NCBI NR]
gi|241879115 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Desulfovibrio vulgaris RCH1 |
| Length: | 704 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNNLYITAT ESKSGKSAVV LGMMQLLLRD VRKVAFFRPI INQASTDVRD HDINLILRHF 60
61 GLDIAYEDTY AYSLQDAREL INNGQHATLL DNILKKYKKL EDAYDFVLCE GTDFLGKDAA 120
121 FEFELNADIA ANLGCPVMVV ANGQQKAARE LIASTQLTID LLDEKGLDVV TAVINRATVT 180
181 EAEREEIIKS LECKVNCSNP LAVYVLPEES TLGKPTMNDV KKWLGAQVLY GHGRLDTLVD 240
241 DYIIAAMQIG NFLDYVSQGC LVITPGDRSD IILSSLASRL STAYPDISGL LLTGGLEPAA 300
301 NVHRLIEGWT GVPIPILSVK DHTYKTIQTL NELYGKIEPD NDRKINTALA LFERHIDSSE 360
361 LGSRLINRKS SRITPKMFEF NLIEKAKRNR MRIVLPEGAE ERILRAADIL VRREVADIIL 420
421 LGDANTVGSR IGDLGLDMDG VQIVQPNLSP KFDEYVAAYH ECRKKKGISM EQARDMMNDP 480
481 TYFGTMMVHK GDADGMVSGA INTTAHTIRP AFEFIKTKPG VSIVSSVFLM CLKDRVLVFG 540
541 DCAVNPNPTA EQLAEIAISA SHTARIFGVD PRVAMLSYST GSSGKGADVE KVIEATRIAK 600
601 ERAPELLLEG PLQYDAAIDM DVARTKLPGS TVAGQATVFI FPDLNTGNNT YKAVQRAAGA 660
661 VAIGPVLQGL NKPVNDLSRG CTVADIVNTV AITAIQAQAE KGLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
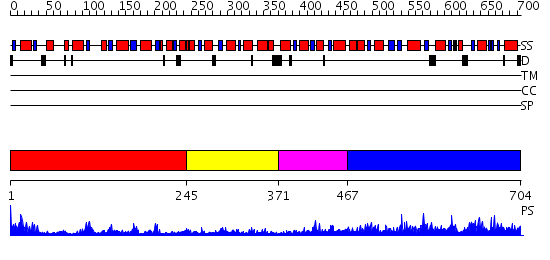
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..244] | 26.221849 | Cell division regulator MinD | |
| 2 | View Details | [245..370] | 2.68 | No description for 2iojA was found. | |
| 3 | View Details | [371..466] | 2.63 | Crystal Structure of a Phosphotransacetylase from Bacillus subtilis | |
| 4 | View Details | [467..704] | 89.522879 | Crystal Structure of a Phosphotransacetylase from Streptococcus pyogenes |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)