













| General Information: |
|
| Name(s) found: |
ORP1C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 814 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
signal transduction
[ISS]
steroid metabolic process [ISS] |
| Molecular Function: |
oxysterol binding
[ISS]
phosphoinositide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHPFCCVTTV SDHSPSMPPL PEPQPPLPNY AADFGSARSE PIITRSASQS YNHSGQFNNN 60
61 LIHSLSFNHH QSDVTDRLGQ RVLALPAAVR EPPVDVKIND IVGNGIAGIL YKWVNYGRGW 120
121 RPRWFVLQDG VLSYYKIHGP DKIFVSPETE KGSKVIGDES ARMISRHNRR GGSSSSCQLR 180
181 RKPFGEVHLK VSSVRESRSD DKRFSIFTGT KRLHLRAETR EDRTTWVEAL QAVKDMFPRM 240
241 SNSELMAPTN NLAMSTEKIR LRLIEEGVSE LAIQDCEQIM KSEFSALQSQ LVLLKQKQWL 300
301 LIDTLRQLET EKVDLENTVV DESQRQADNG CSGELRHEKF SEGTATESDD DNERGDAAEE 360
361 EFDEEENTFF DTRDFLSSSS FKSSGSGFRT SSFSSDEDGF ESEDDIDPSI KSIGCNYPRV 420
421 KRRKNLPDPV EKEKSVSLWS MIKDNIGKDL TKVCLPVYFN EPLSSLQKCF EDLEYSYLLD 480
481 RAFEYGKRGN SLMRILNVAA FAVSGYASTE GRICKPFNPL LGETYEADYP DKGLRFFSEK 540
541 VSHHPMVVAC HCDGTGWKFW GDSNLRSKFW GRSIQLDPVG VLTLQFDDGE ILQWSKVTTS 600
601 IYNLILGKLY CDHYGTMRIE GSAEYSCKLK FKEQSIIDRN PHQVHGIVQN KSGKTVATMF 660
661 GKWDESIHFV TGDCSGKGKL SEDMSGAQLL WKRSKPPGNA TKYNLTRFAI TLNELTPGLK 720
721 ERLPPTDSRL RPDQRYLENG EFEMANTEKL RLEQRQRQAR KMQERGWKPR WFMKEKGSES 780
781 YRYKGGYWEA REDGSWVDCP DIFGHIDSDQ QMIE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
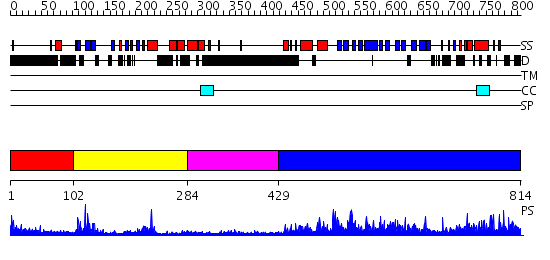
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [102..283] | 13.522879 | Crystal Structure of the PH Domain of SKAP-Hom with 8 Vector-derived N-terminal Residues | |
| 3 | View Details | [284..428] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [429..814] | 70.522879 | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)