













| General Information: |
|
| Name(s) found: |
BAM6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 577 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
carbohydrate metabolic process
[IEA]
polysaccharide catabolic process [IEA] |
| Molecular Function: |
beta-amylase activity
[IEA][ISS]
catalytic activity [IEA] cation binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSVLGMMNP NLINGRNLHK GSSIFVQDKE TKKRVQWRLS IKEGSLRTHQ ATASSATEPK 60
61 ATEFNTTTYE DKMLTNYVPV YVMLQLGVIT NDNVLENEES LKKQLKKLKQ SQVDGVMVDV 120
121 WWGIVESKGP KQYQWSAYRN LFAIVQSFGL KLQAIMSFHR CGGNIGDDVN IPIPKWVLEI 180
181 GDSNPDIFYT NKSGNRNKEC LSLSVDNLSL FRGRTAVEMY RDYMKSFREN MEDFISSGVI 240
241 IDIEVGLGPA GELRYPSYSE TQGWVFPGIG EFQCYDKYLR SDYEEEVRRI GHPEWKLPEN 300
301 AGEYNSVPGE TEFFEYSNGT YLKEEGNFFL SWYSKKLLLH GDQILDEANK VFLGCKLKIA 360
361 AKVSGIHWWY KTESHAAELT AGYYNLKNRD GYRAIAKIMR RHHAILNFTC LEMKNTEQPA 420
421 KAKSGPQELV QQVLSSGWRE GIEVAGENAL PRFDRNGYNQ IILNARPNGV NQDGKPRMFG 480
481 FTYLRLSDKL LNEPNFSTFK MFLKRMHANQ EYCSEPERYN HELLPLERSR NDESLEMFME 540
541 ETEPFDPFPW LDETDMSIRP FESVLSLLRS TFLRKKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
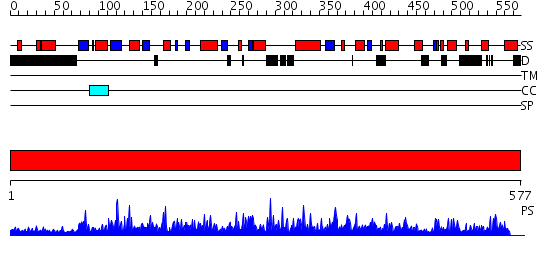
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..577] | 1000.0 | Crystal structure of soybean beta-amylase mutant substituted at surface region |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)