













| General Information: |
|
| Name(s) found: |
ORC1A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
origin recognition complex [ISS] |
| Biological Process: |
DNA replication
[ISS]
regulation of transcription [IMP] |
| Molecular Function: |
DNA binding
[ISS]
double-stranded methylated DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSLSSKAK TFKSPTKTPT KMYRKSYLSP SSTSLTPPQT PETLTPLRRS SRHVSRKINL 60
61 GNDPIDLPGK ESVEEINLIR KPRKRTNDIV VAEKSKKKKI DPEVSFSPVS PIRSETKKTK 120
121 KKKRVYYNKV EFDETEFEIG DDVYVKRTED ANPDEEEEED PEIEDCQICF KSHTNTIMIE 180
181 CDDCLGGFHL NCLKPPLKEV PEGDWICQFC EVKKSGQTLV VVPKPPEGKK LARTMKEKLL 240
241 SSDLWAARIE KLWKEVDDGV YWIRARWYMI PEETVLGRQR HNLKRELYLT NDFADIEMEC 300
301 VLRHCFVKCP KEFSKASNDG DDVFLCEYEY DVHWGSFKRV AELADGDEDS DQEWNGRKEE 360
361 EIDYSDEEIE FDDEESVRGV SKSKRGGANS RKGRFFGLEK VGMKRIPEHV RCHKQSELEK 420
421 AKATLLLATR PKSLPCRSKE MEEITAFIKG SISDDQCLGR CMYIHGVPGT GKTISVLSVM 480
481 KNLKAEVEAG SVSPYCFVEI NGLKLASPEN IYSVIYEGLS GHRVGWKKAL QSLNERFAEG 540
541 KKIGKENEKP CILLIDELDV LVTRNQSVLY NILDWPTKPN SKLVVLGIAN TMDLPEKLLP 600
601 RISSRMGIQR LCFGPYNHRQ LQEIISTRLE GINAFEKTAI EFASRKVAAI SGDARRALEI 660
661 CRRAAEVADY RLKKSNISAK SQLVIMADVE VAIQEMFQAP HIQVMKSVSK LSRIFLTAMV 720
721 HELYKTGMAE TSFDRVATTV SSICLTNGEA FPGWDILLKI GCDLGECRIV LCEPGEKHRL 780
781 QKLQLNFPSD DVAFALKDNK DLPWLANYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
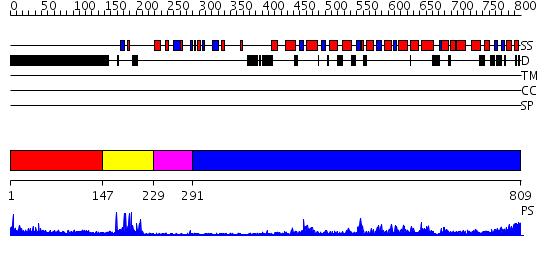
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [147..228] | 13.522879 | No description for 2ysmA was found. | |
| 3 | View Details | [229..290] | 2.81 | Crystal structure of the proximal BAH domain of polybromo | |
| 4 | View Details | [291..809] | 59.0 | The crystal structure of murine p97/VCP at 3.6A |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)