













| General Information: |
|
| Name(s) found: |
C3H24_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
RNA processing
[IEA]
|
| Molecular Function: |
nucleic acid binding
[IEA][ISS]
RNA methyltransferase activity [IEA] zinc ion binding [IEA] methyltransferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METSSIEINE LPLATKTQTP PASVEPIPME TSSIDELPSS DSNATDNIEA VGEKRKRADE 60
61 DEKTNLESSD TKITTPSPWW KTSLCSYFRR EASCSHGNEC KYAHGEAELR MKPDNTWDPT 120
121 SERGKKAKAM KMSEHEEKEE DEVLFTEQMM ESIDGDEGGG GSVSVVDLSL SKCLVHLPNK 180
181 WQSDELKKFL GEQGVLYKSA KKRRGMIVGF VTFENAEQLQ SGVEILDGKT VNSSNLKIAD 240
241 VLPRTFDKND ARKSVKSARD AVTPLAYLSY ADQLEQKKTS IGQMLKKLAR NARKACPNGN 300
301 SLPQWVLTSR DRGGLACNLE GIIESPITNG YRNKCEFSVG LSLQGKPTVG FSLGSFCAGV 360
361 TAVEEPVDCP NVSKIASQYA SIFQKFIENS KFQVWNRFQH SGFWRQLTVR EGRKPGVFSN 420
421 DEDAITRIAE VMLMVQVCLT GSDEAEVATE FEELAKAFAE GARASSPTLP LTVLVVQNHS 480
481 GISNVAPPDA PLQVLAIPIS DNGTDQEQTT NVLTEARIHD HINNLRFSIS PTAFFQVNTV 540
541 TAEKLYSIAG DWADLGPDTL LFDVCCGTGT IGLTLAHRVG MVIGIEMNAS AVADAERNAT 600
601 INGISNCKFI CSKAEDVMSS LLKQYLDVTQ MEEAKPLSNA NDDLNKQIPS TEEMTNSEHV 660
661 ADQNLPPSNT QVEELQDNEQ KDSSSLEPEK TTKPQFKNVV AIVDPPRSGL HPAVIKALRT 720
721 HPRLKRLVYI SCNPETLVAN AIELCTPSFD EPDRGNKNYR GRKKIGIAAL ARHRAKSMPT 780
781 SEAFRPVKAM AVDLFPHTDH CEMVMLLER |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
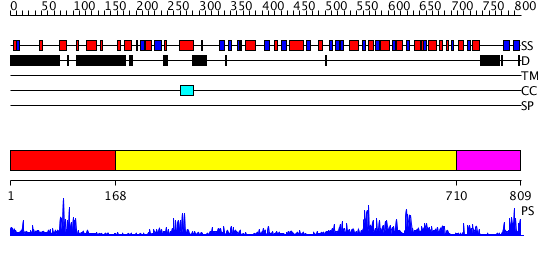
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 14.39794 | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d | |
| 2 | View Details | [168..709] | 65.045757 | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase | |
| 3 | View Details | [710..809] | 20.522879 | Crystal structure of human Dimethyladenosine transferase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)